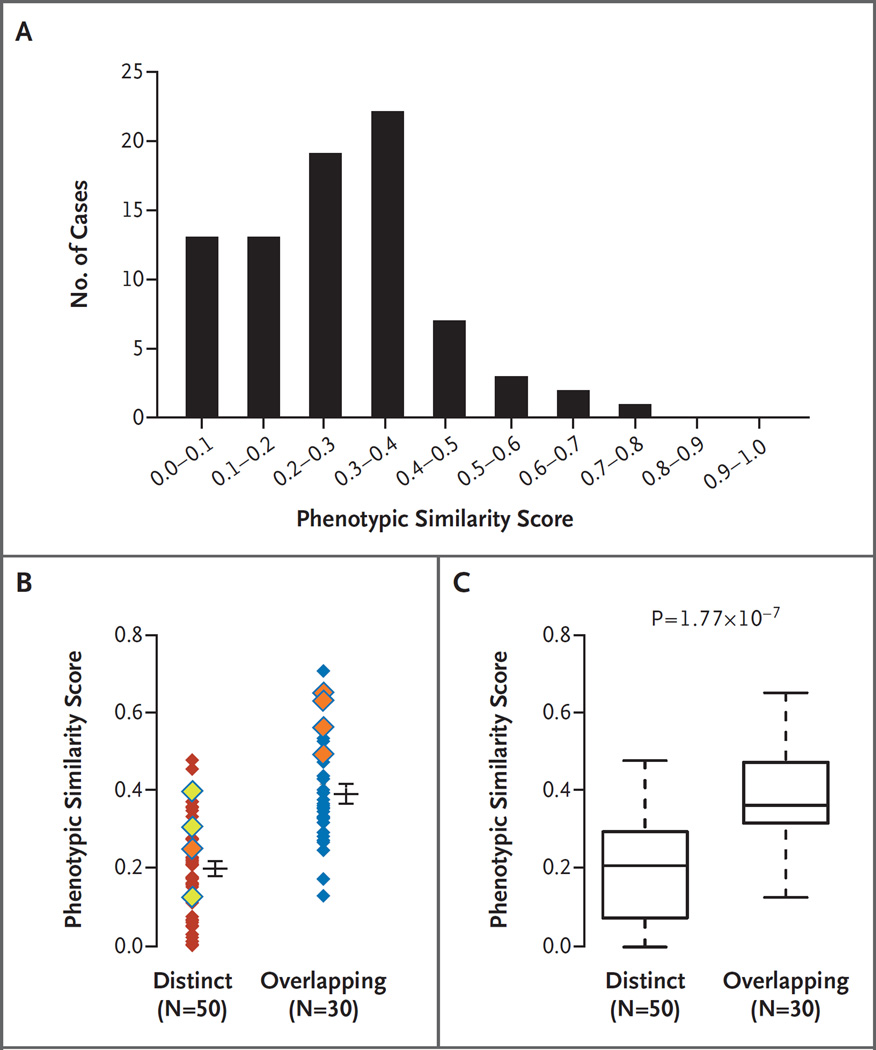
Figure 4. Phenotypic Similarity of 80 Disease Pairs with Blended Phenotypes Categorized as Overlapping or Distinct.
Panel A shows the distribution of cases plotted across the range of phenotypic similarity scores. Panel B shows the relative disease-pair similarity scores calculated by the symmetric Resnik method with the use of Human Phenotype Ontology (HPO) terms assigned to disease identification numbers in the Online Mendelian Inheritance in Man database. Each disease pair is represented by a single diamond; cases classified by two physician scientists as distinct or overlapping are indicated. Increasing phenotypic similarity (determined bioinformatically) corresponds to higher similarity scores. Eight pairs of diseases associated with proteins that have physical or pathway interactions with one another are indicated by yellow diamonds (secondary interaction) or orange diamonds (tertiary interaction); the ninth patient (Patient 12) is not shown here, because neither disease in the pair was mapped to the HPO. Horizontal bars indicate means, and I bars indicate standard errors. Panel C shows box plots of the degree of phenotypic similarity for dual diagnoses that were clinically assigned to distinct or overlapping phenotype categories. The lower and upper limits of the box indicate the first and third quartiles, the bar inside the box the median, and the T bars the minimum and maximum or 1.5 times the interquartile range (whichever value was smaller).