Abstract
The glyoxalase system is a highly specific enzyme system existing in all mammalian cells that is responsible for the detoxification of dicarbonyl species, primarily methylglyoxal (MG). It has been implicated to play an essential role in preventing the increased formation of advanced glycation end products under certain pathological conditions. We have established the first glyoxalase 1 knock-out model (GLO1−/−) in mammalian Schwann cells using the CRISPR/Cas9 technique to investigate compensatory mechanisms. Neither elevated concentrations of MG nor associated protein modifications were observed in GLO1−/− cells. Alternative detoxification of MG in GLO1−/− is achieved by increased catalytic efficiency of aldose reductase toward hemithioacetal (product of glutathione and MG), which is most likely caused by S-nitrosylation of aldose reductase. The hemithioacetal is mainly converted into lactaldehyde, which is paralleled by a loss of reduced glutathione. Inhibition of aldose reductase in GLO1−/− cells is associated with an increased sensitivity against MG, elevated intracellular MG levels, associated modifications, as well as increased oxidative stress. Our data suggest that aldose reductase can compensate for the loss of GLO1. This might be of clinical importance within the context of neuronal diseases caused by an impaired glyoxalase system and elevated levels of dicarbonyl species, such as MG.
Keywords: cell metabolism, CRISPR/Cas, diabetes, enzyme kinetics, glycation, toxicity, aldose reductase, diabetic neuropathy, glyoxalase system, methylglyoxal
Introduction
Advanced glycation end products (AGEs)2 are formed by non-enzymatic glycation of proteins and lipids. AGEs are most likely a risk factor for the progression of late diabetic complications such as diabetic neuropathy (1, 2). Methylglyoxal (MG), a reactive dicarbonyl and major precursor for AGEs, is derived from lipid peroxidation and threonine metabolism but is mainly from glycolysis (3). Hyperglycemic episodes have been linked to increased formation of MG in animals and humans (4–6). MG and associated protein modifications induce cytotoxic effects through the initiation of cell growth arrest, apoptosis, and production of reactive oxygen species (7).
One way to detoxify this reactive metabolite is via the highly specific glyoxalase system, conserved from crude prokaryotics to complex mammalian organisms. The major substrate for the first enzyme in the system, glyoxalase 1 (GLO1), is the non-enzymatically formed product of MG and reduced glutathione (GSH). This spontaneously produced hemithioacetal (HTA) is converted to S-d-lactoylglutathione, which is then hydrolyzed to d-lactate and GSH through the catalytic activity of glyoxalase 2 (8). In comparison with other enzymatic systems involved in the detoxification of endogenous toxins, the glyoxalase system has a limited substrate specificity and is not depleting its cofactor GSH. As the intermediate S-d-lactoylglutathione is a non-toxic metabolite, GLO1 represents the rate-limiting step for the detoxification of MG and also for its toxic capacity (9). It highlights the importance of the glyoxalase system and the common opinion that GLO1 is indispensable to maintain cellular viability (9–11). It has been shown that GLO1 has a reduced capacity in diabetic mouse models and patients, which was paralleled by an accumulation of MG and MG-specific AGEs, particularly in neuronal tissue (4, 12–14). MG-modified proteins in peripheral neurons, like voltage-gated sodium channels, can change the firing patterns of action potentials and consequently the mediation of mechanical triggers like pain in patients suffering from diabetic neuropathy (15). It has been shown previously that sciatic nerves are highly susceptible to the accumulation of MG-associated modifications in diabetes (16). Recently, the critical role of Schwann cell metabolism has been described extensively, and its disruption can cause axonal degeneration. Furthermore, the same study postulated that metabolic dysfunction in the Schwann cell-rich sciatic nerves is associated with the development of neuropathic symptoms in an experimental model of diabetes (17). Schwann cells are critical for the support, development, and regeneration of peripheral neurons. However, there are very few studies that have investigated MG metabolism in these cell types.
Within the context of an alternative detoxification of MG, only some studies have focused on enzymes of the aldehyde dehydrogenase family (ALDH) and aldo-keto reductase family (AKR). Both families have been claimed to convert MG into non-toxic compounds (18). The NADPH-dependent enzymes, such as aldehyde reductase (AKR1a1) and aldose reductase (AKR1b1/AKR1b3) isolated from human placenta and skeletal muscle, are able to convert MG into hydroxyacetone and lactaldehyde (18, 19). In vivo data have confirmed the essential role of AKRs under diabetic conditions in cardiovascular tissue, wherein AKR1b3 null mice had increased AGEs in the heart and showed more atherosclerotic lesion formation (20). ALDHs are NADH-dependent enzymes, and several subtypes (ALDH1, -2, -3, and -9) should be able to convert MG into pyruvate (18, 21). However, the relative contribution of ALDHs to the detoxification of MG in vivo remains unknown. Deglycase DJ-1, also known as Parkinson disease protein 7, can convert MG without GSH directly into lactate in mammalian cells (22). Because of a very low catalytic efficiency as compared with GLO1 (∼1000-fold), the contribution of this enzyme in the context of MG detoxification is uncertain (9, 22). One of the major limitations of these studies is that the efficiency to detoxify MG has been investigated using either purified or recombinant proteins. However, the different expression levels of ALDHs and AKRs in various tissues indicate the difficulty in defining their relative contribution in detoxifying MG in vivo. It is notable that several inhibitors of aldose reductase (AKR1b3) have been used in clinical trials to treat patients suffering from neuropathic symptoms (23, 24). The inhibition of this enzyme to prevent the harmful accumulation of sorbitol in neuronal cells can theoretically have an adverse side effect, given the background of a potentially AKR1b3-driven detoxification of dicarbonyls.
Therefore, the aim of this study was to look at compensatory mechanisms in a neuronal research model system with a GLO1 deficiency. Within this context we expected to show that an alternative enzyme can process dicarbonyl species, and those findings could contribute to improve the molecular understanding of specific neuropathy disorders.
Results
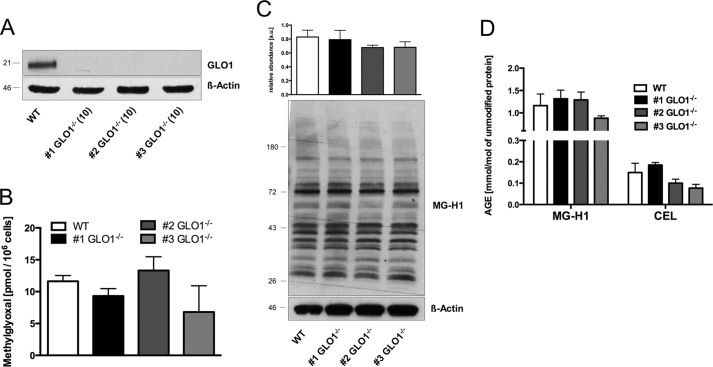
Loss of GLO1 Is Not Associated with Increased Levels of Methylglyoxal or Modified Proteins
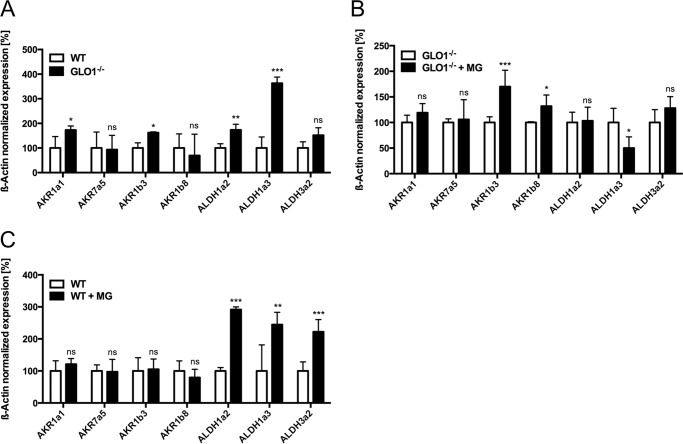
Effective detoxification of MG is mainly accomplished by the glyoxalase system. We established a permanent loss of GLO1 protein and GLO1 activity in murine Schwann cells (Table 1 and Fig. 1A). To investigate the consequences of a GLO1 knock-out under baseline conditions, we determined the levels of MG and MG-specific protein modifications. We detected neither an increase in intracellular MG levels nor MG-associated protein modifications (Fig. 1, B–D). To establish the enzymes potentially involved in the alternative detoxification, we analyzed the mRNA content of 10 AKR and 9 ALDH subtypes in wild-type and GLO1−/− Schwann cells with and without exposure to MG. The results revealed a baseline up-regulation of two subtypes of AKRs (1a1 and 1b3) and two ALDHs (1a2 and 1a3) (Fig. 2A). After the exposure to MG, the mRNA content of AKR1b3 and AKR1b8 was further increased in GLO1−/− Schwann cells (Fig. 2B). In wild-type Schwann cells, the exposure to MG increased the expression of three ALDH subtypes (1a2, 1a3, and 3a2) significantly.
TABLE 1.
GLO1 activity of wild-type Schwann cells and three individual GLO1−/− Schwann cell clones
Activity is described in units, where 1 unit is the amount of GLO1 that catalyzes the formation of 1 μmol of S-d-lactoylglutathione per min. Values are the means of at least three independent experiments ± S.E.
| Clone no. (passage) | GLO1 activity |
|---|---|
| units/mg protein | |
| WT GLO1+/+ (23) | 1.11 ± 0.058 |
| #1, GLO1−/− (10) | 0.005 ± 0.0012 |
| #2, GLO1−/− (10) | 0.009 ± 0.0015 |
| #3, GLO1−/− (10) | 0.008 ± 0.0003 |
FIGURE 1.
Loss of GLO1 is not associated with increased levels of MG or MG-specific AGEs. A, representative Western blotting analysis of total cell extracts (30 μg of protein) from Schwann cells (wild-type and three GLO1−/− clones; in parentheses, passage number after subculturing) probed with anti-GLO1 antibody and anti-β-actin antibody as a loading control. B, intracellular MG levels in wild-type Schwann cells and three individual GLO1−/− Schwann cell clones cultured under baseline conditions (5 mm glucose). C, densitometry analysis and representative Western blotting of total cell extracts (30 μg of protein) from Schwann cells (wild-type and three GLO1−/− clones) probed with anti-MG-H1 antibody detecting MG-modified arginine residues and anti-β-actin antibody as a loading control. D, intracellular AGE levels of MG-modified arginine (MG-H1) and lysine (N-ϵ-(carboxyethyl)lysine; CEL) residues after exhaustive enzymatic digestion and determination via LC-MS/MS. Values are the mean of at least three independent experiments ± S.E. Western blot bar graphs represent the mean of three independent experiments ± S.E.
FIGURE 2.
Several types of oxidoreductases are potentially involved in the detoxification of MG in GLO1−/− Schwann cells. A, baseline mRNA expression of different subtypes of AKR and ALDH in wild-type Schwann cells (□) and three individual GLO1−/− Schwann cell clones (■). Values for wild-type cells are standardized to 100%. B, mRNA expression of different subtypes of AKR and ALDH in three different GLO1−/− Schwann cell clones with (■) and without (□) MG treatment (50 μm; 6 h). C, mRNA expression of different subtypes of AKR and ALDH in wild-type Schwann cells with (■) and without (□) MG treatment (50 μm; 6 h). All data are normalized to β-actin and represent the mean of at least three independent experiments ± S.E. ***, p < 0.0001; **, p < 0.001; *, p < 0.05; and ns, not significant.
S-Nitrosylation of AKR1b3 Is Beneficial for the Efficient Detoxification of Dicarbonyl Species in GLO1−/− Schwann Cells
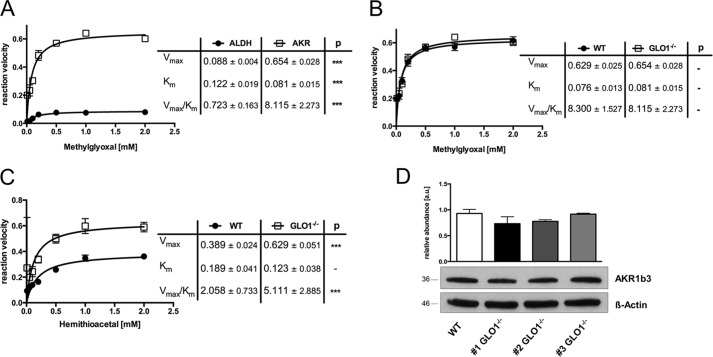
To assess the contribution of the up-regulated enzymes, we determined kinetic profiles for the ALDHs and AKRs present in the cytosolic fractions of GLO1−/− Schwann cells. When MG and the appropriate co-factor (NADPH or NADH) were added as substrate, the Vmax for AKR was 0.654 μmol of NADPH/s/mg of protein, whereas the Vmax for ALDH was only 0.088 NADP/s/mg of protein (Fig. 3A), suggesting that only enzymes of the AKR family are able to detoxify MG efficiently in GLO1−/− Schwann cells. Comparison of AKR kinetics with wild-type Schwann cells revealed the same efficiency in catalyzing the conversion of MG (Fig. 3B). When HTA (non-enzymatic product of MG and GSH) was used in the same enzymatic assay as a substrate, the capacity and affinity were increased in GLO1−/− Schwann cells as compared with wild-type Schwann cells (Fig. 3C). This observation was not caused by an increased amount of the most abundant AKR subtype AKR1b3 in GLO1−/− Schwann cells (Fig. 3D).
FIGURE 3.
Aldo-keto reductases contribute to the efficient detoxification of hemithioacetal. A, kinetic profile of the AKR- (□) and ALDH (●)-catalyzed reduction of MG in the cytosol of GLO1−/− Schwann cells. B, kinetic profile of the AKR-catalyzed reduction of MG in wild-type (●) and GLO1−/− (□) Schwann cells. C, kinetic profile of the AKR-catalyzed reduction of HTA in wild-type (●) Schwann cells and three individual GLO1−/− (□) Schwann cell clones. AKR, Vmax (μmol of NADPH/min/mg protein); Km (mm); ALDH, Vmax (μmol of NADH/min/mg protein); Km (mm). D, densitometry analysis and representative Western blot of total cell extracts (30 μg of protein) from Schwann cells (wild-type and three GLO1−/− clones) probed with anti-AKR1b3 antibody and anti-β-actin antibody as a loading control. All kinetic data represent the mean of at least four independent experiments ± S.E. Western blot bar graphs represent the mean of three independent experiments ± S.E. ***, p < 0.0001.
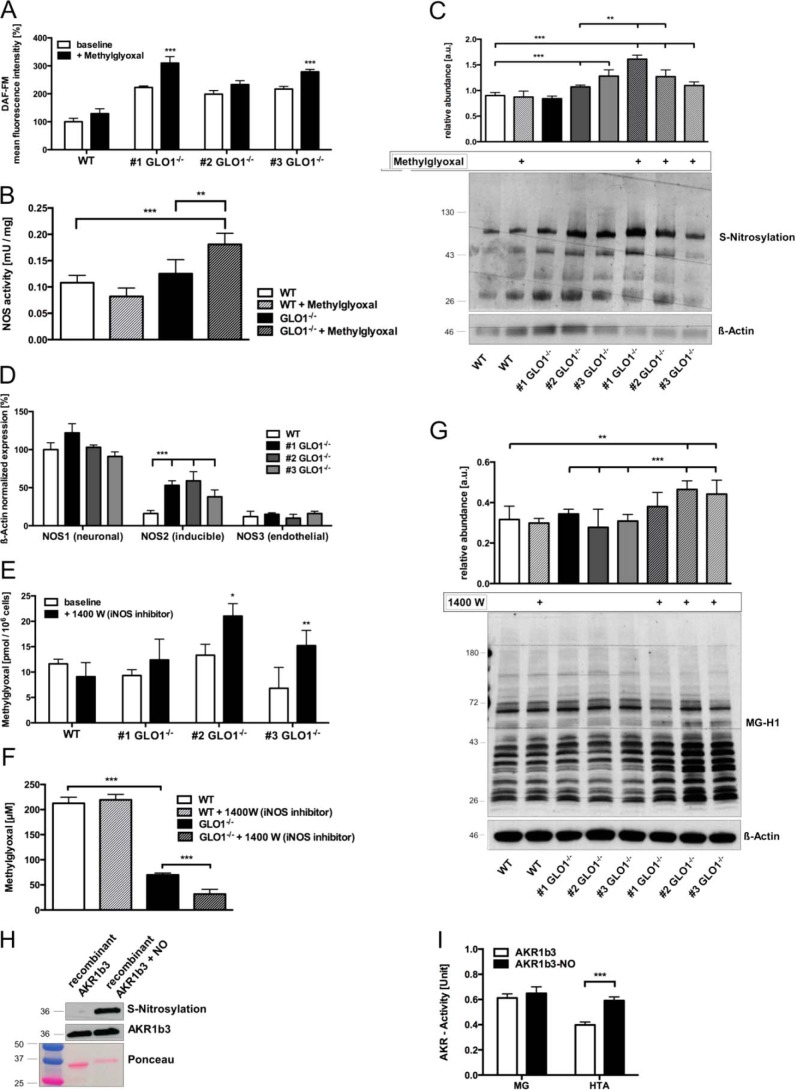
To investigate the reason for the increased catalytic activity in GLO1−/− Schwann cells toward the substrate HTA, we detected higher intracellular levels of nitric oxide (NO) and higher amounts of nitrosylated cysteine residues in GLO1−/− Schwann cells (Fig. 4, A and C). Analysis of the mRNA expression of all NOS subtypes revealed NOS2 or iNOS as the driving force for increased nitric oxide species in GLO1−/− Schwann cells (Fig. 4D). The specific inhibition of this isoform was associated with increased MG levels and MG-modified proteins in two of the GLO1−/− Schwann cell clones (Fig. 4, E and G) and also with a decreased tolerance against exogenously added MG in a toxicity assay (Fig. 4F). Additionally, we found that the exposure to relatively low MG concentrations (50 μm) was accompanied by increased levels of NO, NOS activity, and S-nitrosylation in GLO1−/− Schwann cells (Fig. 4, A–C). The incubation of recombinant AKR1b3 with an NO donor (streptozotocin) revealed an ∼2-fold increased activity toward HTA as a substrate as compared with the unmodified recombinant AKR1b3 (Fig. 4, H and I).
FIGURE 4.
S-Nitrosylation of AKR1b3 is beneficial for the efficient detoxification of dicarbonyl species in GLO1−/− Schwann cells. A, intracellular levels of nitric oxide species in wild-type (□) Schwann cells and three individual GLO1−/− (■) Schwann cell clones using flow cytometry and DAF-FM as a dye reagent. B, enzymatic activity of nitric-oxide synthases in wild-type and GLO1−/− Schwann cells (clone 1), where 1 unit of NOS activity is the amount of enzyme required to yield 1 μmol of nitric oxide/min. C, densitometry analysis and representative Western blot of whole cell lysates of wild-type Schwann cells and three individual GLO1−/− Schwann cell clones with and without MG treatment (50 μm; 6 h) detecting nitrosylated cysteine residues using the iodoTMT switch technique. Lysates were probed with anti-iodoTMT and β-actin as a loading control. D, baseline mRNA expression of the three different subtypes of nitric-oxide synthases (NOS) in wild-type (□) Schwann cells and three individual GLO1−/− (■) Schwann cell clones. Values for NOS1 in wild-type Schwann cells were standardized to 100%. E, intracellular MG levels in wild-type Schwann cells and three individual GLO1−/− Schwann cell clones treated with iNOS inhibitor 1400 W dihydrochloride (100 μm; 24 h). F, median lethal doses (LD50) for MG (48 h exposure time) in wild-type and GLO1−/− Schwann cells (clone 1) treated with iNOS inhibitor 1400 W dihydrochloride (100 μm). G, densitometry analysis and representative Western blot of total cell extracts (30 μg of protein) from wild-type Schwann cells and three individual GLO1−/− Schwann cell clones with and without 1400 W treatment (100 μm; 24 h), probed with anti-MG-H1 antibody detecting MG-modified arginine residues and anti-β-actin antibody as a loading control. H, densitometry analysis and representative Western blot of recombinant AKR1b3 protein with and without exposure to a nitric oxide donor (streptozotocin). Blots were probed with anti-AKR1b3 antibody and stained with Ponceau S staining as a loading control. I, enzymatic activity assay of recombinant AKR1b3 under substrate saturating conditions (2 mm of each) with (■) and without (□) the exposure to a NO donor, where 1 unit is the amount of AKR1b3 that catalyzes the formation of 1 μmol of NADP in 1 min. All data represent the mean of at least four independent experiments ± S.E. Western blot bar graphs represent the mean of three independent experiments ± S.E. ***, p < 0.0001; **, p < 0.001; *, p < 0.05.
AKR1b3 Has an Essential Role in Maintaining Cellular Viability and Preventing Accumulation/Formation of MG and MG-modified Proteins and Oxygen Species
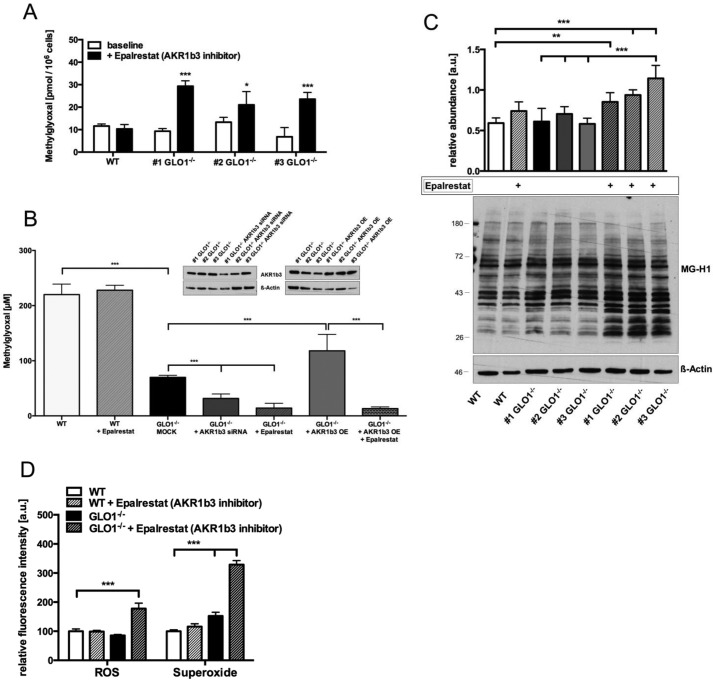
To investigate the contribution of AKR1b3, we determined the median lethal dose (LD50) for MG under different conditions. Wild-type Schwann cells had a baseline LD50 of 220 ± 19 μm MG regardless of whether they had been treated with an AKR1b3 inhibitor (epalrestat) (Fig. 5B). In three individual GLO1−/− Schwann cell clones, the baseline tolerance against MG was decreased to an LD50 of 69 ± 3.7 μm MG (Fig. 5B). The treatment with an AKR1b3 inhibitor (epalrestat) or the usage of siRNA against AKR1b3 was accompanied by almost a total loss of MG tolerance (AKR1b3 siRNA, LD50 = 31.6 ± 9.2; epalrestat, LD50 = 14.2 ± 8.6) (Fig. 5B). The overexpression of AKR1b3 in three individual GLO1−/− Schwann cell clones could prevent the decreased tolerance toward MG (AKR1b3 overexpression, LD50 = 118.1 ± 29.8 μm), whereas the treatment of epalrestat similarly diminished this rescue effect (Fig. 5B).
FIGURE 5.
AKR1b3 has an essential role in maintaining cellular viability and preventing MG accumulation, formation of MG-modified proteins, and oxygen species. A, intracellular MG levels in wild-type Schwann cells and three individual GLO1−/− Schwann cell clones treated with AKR1b3 inhibitor epalrestat (10 μm; 24 h). B, median lethal doses (LD50) for MG (48-h exposure time) in wild-type Schwann cells and three individual GLO1−/− Schwann cell clones treated with AKR1b3 inhibitor epalrestat (10 μm), transfected with siRNA against AKR1b3 or control siRNA (MOCK), and transfected with mammalian expression vector for AKR1b3 (overexpression, OE). Representative Western blots probed with anti-AKR1b3 antibody and anti-β-actin antibody as a loading control demonstrate the decrease (siRNA) or increase (overexpression) in AKR1b3 protein expression. All data represent the mean of at least six independent experiments ± S.E. C, densitometry analysis and representative Western blot of total cell extracts (30 μg of protein) from wild-type Schwann cells and three individual GLO1−/− Schwann cell clones with and without epalrestat treatment (10 μm; 24 h), probed with anti-MG-H1 antibody detecting MG-modified arginine residues and anti-β-actin antibody as a loading control. D, intracellular levels of H2O2 or superoxide species in wild-type (WT) and GLO1−/− Schwann cells (clone 1) treated with AKR1b3 inhibitor epalrestat (10 μm; 24 h) as determined by flow cytometry. ROS, reactive oxygen species. All data represent the mean of at least four independent experiments ± S.E. Western blot bar graphs represent the mean of three independent experiments ± S.E. ***, p < 0.0001; **, p < 0.001; *, p < 0.05.
The inhibition of AKR1b3 with epalrestat was associated with significantly increased levels of MG and MG-modified arginine residues in all three GLO1−/− Schwann cell clones (Fig. 5, A and C). Additionally, we found elevated levels of hydrogen peroxide (2-fold) and superoxide species (3-fold) following AKR1b3 inhibition with epalrestat in GLO1−/− Schwann cells (Fig. 5D).
GSH Reductase Is Required to Preserve Cellular GSH Concentration in GLO1−/− Schwann Cells and to Assist in the Detoxification of Dicarbonyl Species
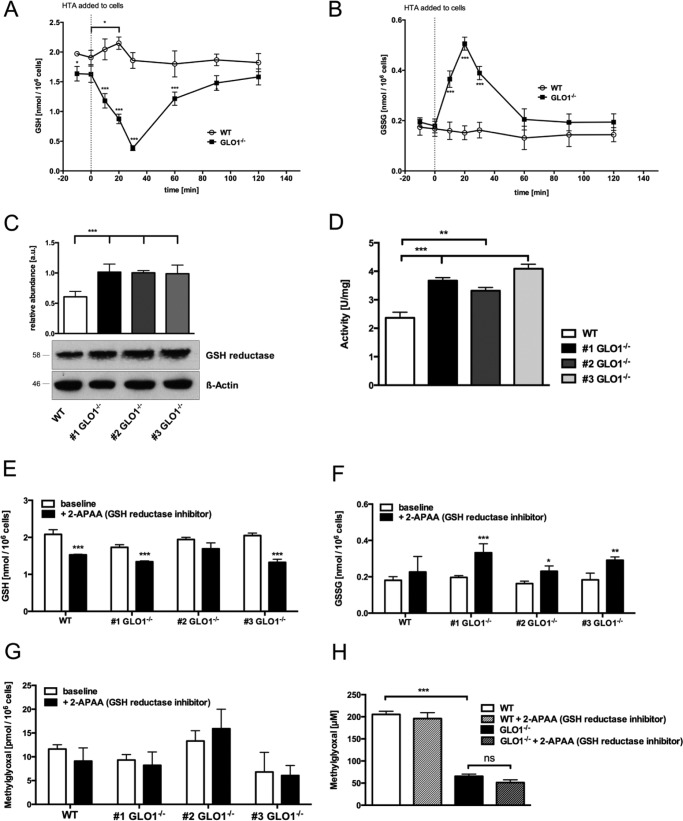
As GSH is a fundamental cofactor in the context of MG detoxification, its fate after the conversion of HTA in wild-type and GLO1−/− Schwann cells was investigated. Both cell lines were exposed to HTA, and the GSH/GSSG content was determined at different time points. In the wild-type Schwann cells a slight increase of GSH after 20 min was detectable, whereas the GSSG content showed no change over the whole time period (Fig. 6, A and B). In three individual GLO1−/− Schwann cell clones, GSH was rapidly diminished after the exposure to HTA. This was paralleled by an increase in GSSG, which was equalized 60 min after the exposure with HTA (Fig. 6, A and B).
FIGURE 6.
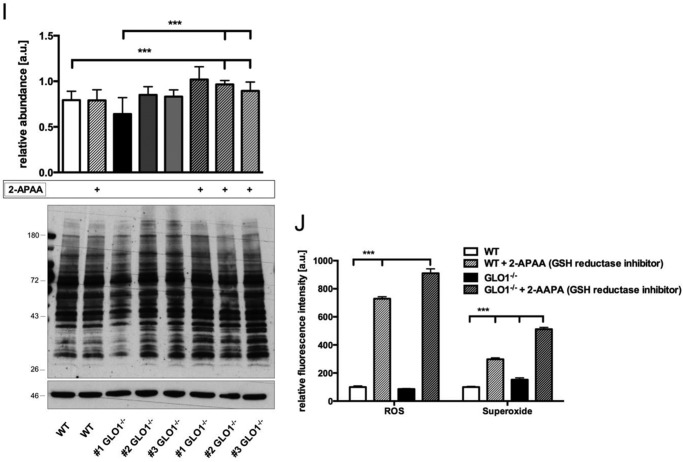
GSH reductase is required to preserve cellular GSH concentration in GLO1−/− Schwann cells and to assist in the detoxification of dicarbonyls. A, cytosolic GSH content after the incubation of wild-type (○) Schwann cells or three individual GLO1−/− (■) Schwann cell clones with HTA (0.2 mm) for 10, 20, 30, 60, 90, and 120 min. The dotted line indicates the time point (0 min) when HTA was added to the cells. B, cytosolic GSSG content after the incubation of wild-type (○) Schwann cells or three individual GLO1−/− (■) Schwann cell clones with HTA (0.2 mm) for 10, 20, 30, 60, 90, and 120 min. The dotted line indicates the time point (0 min) when HTA was added to the cells. All data represent the mean of at least three independent experiments ± S.E. C, densitometry analysis and representative Western blot of total cell extracts (30 μg of protein) from Schwann cells (wild-type and three GLO1−/− clones) probed with anti-GSH reductase antibody and anti-β-actin antibody as a loading control. D, enzymatic activity assay of GSH reductase in the cytosol of wild-type (□) Schwann cells or three individual GLO1−/− (■) Schwann cell clones under substrate saturating conditions (5 mm GSSG), where 1 unit is the amount of GSH reductase that catalyzes the formation of 1 μmol of NADP in 1 min. E, cytosolic GSH content in wild-type (□) Schwann cells or three individual GLO1−/− (■) Schwann cell clones treated with GSH reductase inhibitor 2-APAA (5μmm, 6 h). F, cytosolic GSSG content in wild-type (□) Schwann cells or three individual GLO1−/− (■) Schwann cell clones treated with GSH reductase inhibitor 2-APAA (5 μm, 6 h). G, intracellular MG levels in wild-type (□) Schwann cells and three individual GLO1−/− (■) Schwann cell clones treated with GSH reductase inhibitor 2-APAA (5 μm; 24 h). H, median lethal doses (LD50) for MG (48 h exposure time) in wild-type and GLO1−/− Schwann cells (clone 1) treated with GSH reductase inhibitor 2-APAA (5 μm). I, densitometry analysis and representative Western blot of total cell extracts (30 μg of protein) from wild-type Schwann cells and three individual GLO1−/− Schwann cell clones with and without 2-APAA treatment (5 μm; 24 h), probed with anti-MG-H1 antibody detecting MG-modified arginine residues and anti-β-actin antibody as a loading control. J, intracellular levels of H2O2 or superoxide species in wild-type and GLO1−/− Schwann cells (clone 1) treated with GSH reductase inhibitor 2-APAA (10 μm; 24 h) as determined by flow cytometry. ROS, reactive oxygen species. All data represent the mean of at least four independent experiments ± S.E. Western blot bar graphs represent the mean of three independent experiments ± S.E. ***, p < 0.0001; **, p < 0.001; *, p < 0.05; ns, not significant.
Analysis of the activity and the protein content of GSH reductase revealed an ∼2-fold increase of both in GLO1−/− Schwann cells (Fig. 6, C and D). In wild-type and GLO1−/− Schwann cells, the inhibition of GSH reductase was paralleled with a decrease in GSH (Fig. 6E). Only in GLO1−/− Schwann cells was this inhibition associated with enhanced GSSG levels (Fig. 6F).
However, the inhibition of GSH reductase was not associated with a decreased tolerance toward MG in GLO1−/− Schwann cells (Fig. 6H). Within the context of MG metabolism, this inhibition of GSH reductase led only to a minor and non-significant increase in MG but to a significant increase in MG-modified proteins in three individual GLO1−/− Schwann cells (Fig. 6, G and I). Additionally, the inhibition of GSH reductase was accompanied by an increase of both oxygen species in wild-type and GLO1−/− Schwann cells (Fig. 6J).
Products of Alternative Detoxification Are Hydroxyacetone and Lactaldehyde
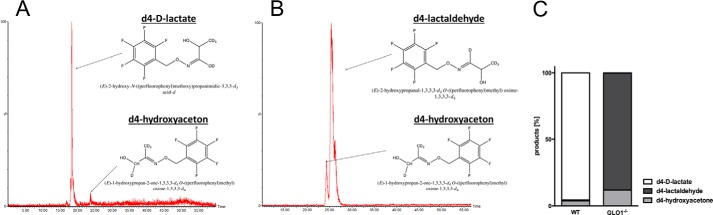
To determine the products of this alternative detoxification pathway, labeled d4-MG was used. In wild-type Schwann cells the most abundant peak was d4-d-lactate paralleled by a slight increase in d4-hydroxyacetone (Fig. 7, A and C). Incubation of cytosolic fractions from GLO1−/− Schwann cells with d4-MG revealed no abundance of d4-d-lactate but an increase in d4-hydroxyacetone and d4-lactaldehyde (Fig. 7B). The distribution for those products was in favor of lactaldehyde (90%) (Fig. 7C).
FIGURE 7.
Product distribution after in vivo conversion of methylglyoxal in GLO1−/− Schwann cells is in the favor of lactaldehyde. A, representative chromatogram of intracellular d4 products in wild-type Schwann cells 60 min after the exposure to d4-MG. Chemical structure displays the complex of labeled d4-d-lactate/d4-hydroxyacetone and the derivatizing agent (PFBHA) B, representative chromatogram of intracellular d4 products in GLO1−/− Schwann cells 60 min after the exposure to d4-MG. Chemical structure displays the complex of labeled d4-lactaldehyde/d4-hydroxyacetone and the derivatizing agent (PFBHA) C, product distribution of isotopically labeled intracellular metabolites in percent after the conversion of d4-MG in vivo. Wild-type and GLO1−/− Schwann cells were exposed to isotopically labeled d4-MG (100 μm) for 60 min, and products were analyzed by LC-MS/MS.
Discussion
Schwann cells are responsible for many aspects of peripheral nerve biology, such as maintenance of axonal integrity, insulation of axons, localization of sodium channels, and nerve regeneration. In diabetic neuropathy, most of those aspects are malfunctioning (25, 26). Increased oxidative stress caused by reactive compounds, derived from a disturbed energy metabolism, contributes to the dysfunction of Schwann cells. The inefficient detoxification of those reactive compounds, such as MG, may indirectly be linked to the progression of diabetic neuropathy (27).
The glyoxalase system has been shown to have a key role in preventing the harmful events of glycated proteins and lipids via the removal of dicarbonyl species (7, 9, 28). The knockdown of GLO1 was associated with diabetic nephropathy in healthy wild-type mice, and a decreased capacity of GLO1 resulted in dramatic accumulation of MG in diabetic patients (6, 29). In contrast, the overexpression of GLO1 could prevent the accumulation of MG and AGEs and has shown cyto-protective effects in vitro and in vivo (10, 30). Recent research has focused on the development of inducers for GLO1 activity, and the first clinical trials have taken place for the establishment of a GLO1 enhancer in diabetic patients (31, 32). The attention paid to this topic therefore highlights the importance of GLO1 in preventing the progression of diabetic complications.
Surprisingly, in our GLO1−/− model system we detected neither elevated MG nor MG-specific protein modifications. This can be explained by a highly efficient AKR1b3, which tries to imitate GLO1 in catalyzing excessive amounts of HTA, the spontaneous product of MG and GSH. The higher catalytic efficiency of GLO1−/− Schwann cells in converting HTA is likely due to a post-translational modification of AKR1b3. The up-regulation of iNOS and the increased NO levels are paralleled by elevated amounts of nitrosylated cysteine residues. Interestingly, it has been shown frequently that MG can impair NO homeostasis, and this has been related to endothelial dysfunction via a modulation of endothelial NOS (33, 34). Chang et al. (35) concluded that the increased generation of NO in endothelial cells was also related to the production of peroxynitrite and therefore to the harmful effects of NO. In our study, recombinant AKR1b3, which has been nitrosylated ex vivo, showed the same increased kinetic efficiency toward HTA similar to the GLO1−/− Schwann cells. It was also shown that exogenous MG increased the intracellular levels of NO even further, and in turn the inhibition of iNOS was associated with an impaired MG metabolism. Given this background, we have strong indications for a NOS-driven post-translational modification of AKR1b3, which leads to a more efficient catalysis of GSH-bound MG (HTA). Indeed, the S-nitrosylation of AKR1b3 at Cys-298 and Cys-303 has been described elsewhere and is associated with an increased activity of the enzyme that can be reversed by glutathiolation (36, 37). Within this context, it is interesting to note that the conversion of HTA by AKR1b3 in GLO1−/− Schwann cells was paralleled by an intracellular change in the ratio of oxidized/reduced GSH, potentially caused by the necessity for a glutathiolation of nitrosylated AKR1b3 to reverse it into a reduced form. The inhibition of GSH reductase showed that GSSG levels are rising rapidly in GLO1−/− Schwann cells. It seems to play a crucial role in those cells to maintain the intracellular GSH pool in a range to prevent cell death.
However, GLO1−/− Schwann cells would have an increased demand for reducing agents like NADPH. This is on the one side a result of the alternative detoxification through AKR1b3, and on the other side it is due to the reduction of oxidized GSH. Generation of reducing agents is only achievable with an increased glycolysis, particularly with the pentose phosphate pathway. Consequently, this would lead to increased reactive oxygen species and MG production that causes the necessity for even more reducing agents like NADPH, leading to a vicious circle.
Vander Jagt et al. (18, 38) had previously shown that AKR1b3 can act as an aldehyde-reducing enzyme or a ketone-reducing enzyme, resulting in the production of hydroxyacetone and lactaldehyde, respectively. In our study, we confirmed the data in vivo using isotopically labeled d4-MG. The existence of ∼2% d4-hydroxyacetone in wild-type Schwann cells after the conversion of d4-MG indicates that the contribution of alternative detoxification through AKR1b3 is minimal in healthy wild-type Schwann cells. In GLO1−/− Schwann cells, the main product is lactaldehyde due to a change in substrate specificity of AKR1b3. Previous studies that have focused on alternative enzymes for the detoxification of MG were based upon the idea that free unhydrated MG will be detoxified directly through alternative enzymes into hydroxyacetone or pyruvate, depending on the tissue distribution of AKRs and ALDHs (18, 21). However, the intracellular GSH concentrations are kept in a range that is 100-fold higher than cytosolic MG concentrations (38). This guarantees an immediate binding of MG to GSH to form the substrate of GLO1. As GSH is present in excessive amounts in the cytosol and HTA has a dissociation constant of ∼3 mm, free MG can only exist in low nanomolar traces in the cytosol, suggesting its concentration is below the lowest affinity of AKRs and ALDHs (39–41).
AKR1b3, commonly known as aldose reductase, is a ubiquitously expressed enzyme, especially in cells of the kidney and the peripheral nervous system. It is a catalyst with a broad range of substrates with physiological significance, such as 4-hydroxynonenal, isocorticosteroids, glutathione conjugates, or pollutants like acrolein (42). Much attention has been paid to this protein for its crucial role as the initial enzyme of the polyol pathway, where it produces sorbitol during hyperglycemic episodes in various tissues (25, 43). Glucose is preferentially taken up into Schwann cells in peripheral neurons, which have naturally high expression levels of AKR1b3 compared with cells of other tissues (44, 45). This is the reason why a lot of effort has been made in the last 2 decades to develop AKR1b3 inhibitors to prevent diabetic complications caused by high sorbitol levels (23). However, the results of a large number of new therapies tested in clinical trials and animal experiments are contradictory. From nine AKR1b3 inhibitors only epalrestat showed slightly beneficial and non-adversarial health effects, and it is only marketed for usage in Japan and India and not in the United States or the European Union (24, 46). In this study, we have demonstrated that the inhibition of AKR1b3 by epalrestat, when GLO1 is lost, is associated with increased MG concentration and elevated AGE levels. It is therefore conceivable that in diabetes the loss of GLO1 activity leads to the detoxification of MG by AKR1b3. Within this context, the inhibition of AKR1b3, far from preventing the accumulation of sorbitol, blocks the remaining pathway by which other reactive metabolites can be detoxified, leading to their accumulation and increased post-translational modifications. The double-edged role of AKR1b3, in activating the polyol pathway on the one side and detoxifying reactive metabolites on the other side, is paradoxical and has to be investigated in further studies. However, our study provides a mechanism to explain why several AKR1b3 inhibitors have failed in clinical trials for the treatment of diabetic complications. It shows for the first time a way to survive dicarbonyl-mediated stress without the glyoxalase system in mammalian cells. Although the cells are highly susceptible to exogenously added MG, the intracellular MG concentrations of GLO1−/− Schwann cells are held in a range similar to wild-type cells. Undoubtedly, AKR1b3 contributes to the detoxification of AGE precursors in cells of the peripheral nervous system, whereas GLO1 is not as efficient as it should be. Within a clinical context where GLO1 activity is reduced, such as in diabetic and elderly patients, there is no clear-cut relationship between patient outcome and GLO1 activity. This fact potentially indicates the existence of an alternative pathway for MG detoxification or the possibility that a low expression of GLO1 is sufficient.
Experimental Procedures
Cell Culture
Primary murine Schwann cells immortalized with SV40 large T antigen were obtained from the ATCC® (Manassas, VA). Cells were grown in DMEM (Gibco) with 1 g/ml glucose containing 10% FCS (Sigma), 1% penicillin (10,000 units/ml) (Gibco), 1% streptomycin (10 mg/ml) (Gibco), and 1% amphotericin B (250 μg/ml) (Gibco) at 33 °C in a saturated humidity atmosphere containing 95% air and 5% CO2. Cells were grown to 70% confluence for in vivo experiments and passaged at 90% confluence using 0.05% trypsin/EDTA (Gibco).
Synthesized Chemicals
MG solution (40% w/v) was purchased from Sigma. d4-MG was synthesized, as described previously (47), using d6-acetone. The concentration of MG was determined by LC-MS/MS, as described. The concentration and purity of the stock solutions were determined by 13C and 1H NMR using a Bruker AVANCE II NMR spectrometer. The total purity of the unlabeled and labeled MG solutions was 60–65% with the major contaminants being methanol, acetate, and acetone.
Antibody against MG-H1
A rat monoclonal antibody against MG-H1 was developed in collaboration with the monoclonal antibody core facility at Helmholtz Centre in Munich, Germany. Briefly, a peptide-based antigen was synthesized that contained a single chemically defined MG-H1 residue (48). Following immunization of LouC rats with ovalbumin-coupled MG-H1-modified peptide, hybridomas were generated according to standard procedures. Hybridomas were screened for reactivity toward the MG-H1 peptide antigen, and human serum albumin was modified minimally with MG by ELISA and Western blotting (49). Positive wells were established and cloned twice by limiting dilution. One anti-MG-H1 monoclonal antibody recognizing MG-H1, designated 6D7 (IgG2c), was used in this study as culture supernatant. A more detailed characterization of the anti-MG-H1 antibody will be published elsewhere.
Generation of CRISPR/Cas9-induced Glo1 Knock-out Cells
Four sgRNAs (76, 171, 183, and 187) targeting exon1 and -2 of the Glo1 locus were designed and cloned into a U6 promoter-driven expression vector (pU6-gRNA) by Sigma. All sgRNAs were tested for efficiency by transfecting together with the Cas9-expressing plasmid pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene plasmid ID 42230) into C3H/10T1/2 mouse embryonic cells. After 72 h, DNA was extracted using a Qiagen QIAamp® DNA mini kit. T7 endonuclease assay using T7 endonuclease I (New England Biolabs) was performed with primers flanking the modified site. sgRNA171 revealed the most efficient performance with a gene editing efficiency of 37% (specific recognition site, 5′-AGCACCAAGGTGGGTGAC-3′). GLO1-deficient Schwann cells were established by transfecting with sgRNA171, plasmid pX330, and mTurquoise-expressing plasmid (the kind gift from Dr. Joachim Goedhart (50)) using a NEON® electroporation transfection system (Thermo Fisher Scientific) with the following conditions: pulse voltage, 1300 mV; pulse width, 20 ms; pulse number, 2. Schwann cells were either transfected with an empty plasmid containing only an mTurquoise vector (wild type) or with the plasmid containing mTurquoise, sgRNA_171 and pX330 (GLO1−/−). Transfected cells were then sorted as single cells by FACSAriaII (BD Biosciences). Positive transfected cells were detected by ultraviolet laser (excitation, 355 nm; filter, 379/28 nm) and were then cultured as sorted single cells and grown to 80% confluence before subculturing. In total, 19 clones based on a single cell seeding strategy were cultured and analyzed for GLO1 activity and protein expression. Based upon sequencing analysis, clones 1, 3, and 4 were chosen for experiments, which were then renamed as 1, 2, and 3, see under “Results.” The methods and the sequencing results for those three individual clones are summarized in the supplemental Materials and Methods.
Overexpression/Silencing of AKR1b3 in GLO1−/−Schwann Cells
Host Escherichia coli strain DH10B, including a mammalian expression vector (pCMV-SPORT6) for AKR1b3 (GE Healthcare; clone ID, 3582788; insert sequence, BC004725), was plated onto LB plates, including 100 μg/ml ampicillin, and incubated overnight at 37 °C. Three individual clones were picked and amplified in LB broth, including antibiotics for 12 h at 37 °C, and isolated using GenEluteTM HP Plasmid MaxiPrep kit (Sigma). Integrity of purified expression constructs was validated by gel electrophoresis, and the concentration was determined by absorbance measurements. 1 × 106 GLO1−/− Schwann cells were transfected with 20 μg of AKR1b3 vector using a NEON® electroporation transfection system (Thermo Fisher Scientific) with the conditions described above. Specific AKR1b3 down-regulation/silencing was achieved using siGENOME AKR1b3 siRNA (GE Healthcare; RNA accession number NM_009658.3; protein accession number NP_033788.3). Control groups (MOCK siRNA) were transfected with siGENOME non-targeting siRNA pool (GE Healthcare; target firefly luciferase mRNA U47296). 1 × 106 GLO1−/− Schwann cells were transfected with 500 pmol of siRNA using a NEON® electroporation transfection system (Thermo Fisher Scientific) with the conditions described above. For both transfection approaches, the cells were allowed to acclimatize for 6 h and were then used for further experiments. Efficiency was assessed by Western blotting 24 h after treatment.
Quantitative PCR
Extraction of RNA was achieved using a peqGOLD MicroSpin Total RNA kit (Peqlab), which was then converted into cDNA with a high capacity cDNA reverse transcription kit (Thermo Fisher Scientific). qPCR was performed using DyNAmo ColorFlash SYBR Green qPCR Master Mix (Thermo Fisher Scientific) and a LightCycler® 480 Instrument II (Roche Applied Science). Signals of amplified products were verified using melting curve analysis, and mRNA levels were normalized to β-actin. Relative expression levels were calculated using the ΔΔCt method described elsewhere (51). Primer sequences used for analyzing mRNA content were as follows: AKR1a1 (PrimerBank ID 10946870a1), forward 5′-AGCCTGGTCAGGTGAAAGC-3′ and reverse 5′-GGCCTCCCCAATCTCAGTT-3′; AKR1b3 (PrimerBank ID 31981909a1), forward 5′-AGGCCGTGAAAGTTGCTATTG-3′ and reverse 5′-ATGCTCTTGTCATGGAACGTG-3′; AKR1b8 (PrimerBank ID 6679791a1), forward 5′-GACCAAGGCAGAATCCTCACC-3′ and reverse 5′-AGATGCCCTTCGAGTGACAGT-3′; AKR1cl (PrimerBank ID 13386240a1), forward 5′-TTGGAACAATCCCTGAGAAAGC-3′ and reverse 5′-TGGCTAACCCTGAATCCTTACA-3′; AKR7a5 (PrimerBank ID 27659728a1), forward 5′-CGGCCAGTCCGAGAACATC-3′ and reverse 5′-TTCAGTGACTTCCCTTCCCAG-3′; ALDH1a2 (PrimerBank ID 6677665a1), forward 5′-CAGAGAGTGGGAGAGTGTTCC-3′ and reverse 5′-CACACAGAACCAAGAGAGAAGG-3′; ALDH1a3 (PrimerBank ID 31542123a1), forward 5′-GGGTCACACTGGAGCTAGGA-3′ and reverse 5′-CTGGCCTCTTCTTGGCGAA-3′; ALDH3a2 (PrimerBank ID 6680678a1), forward 5′-TCTCTGCCCTTTGGAGGTGT-3′ and reverse 5′-AGCTGATCCTTGACAATCACAG-3′; NOS1 (PrimerBank ID 6724321a1), forward 5′-CTGGTAAGGAACGGGTCAG-3′ and reverse 5′-CCGATCATTGACGGCGAGAAT-3′; NOS2 (PrimerBank ID 6754872a1), forward 5′-GTTCTCAGCCCAACAATACAAGA-3′ and reverse 5′-GTGGACGGGTCGATGTCAC-3′; NOS3 (PrimerBank ID 31982150a1), forward 5′-GGCTGGGTTTAGGGCTGTG-3′ and reverse 5′-CTGAGGGTGCGTAGGTGATG-3′.
Preparation of Total/Cytosolic Protein Extracts
For cytosolic extracts 500 μl of cold lysis buffer (10 mm HEPES, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 0.05% Nonidet P-40 supplemented with a pre-made protease inhibitor mixture (Sigma), including 4-(2-aminoethyl)benzenesulfonyl fluoride, aprotinin, bestatin, E-64, leupeptin, pepstatin A) was added to 3 × 106 cells. Samples were then homogenized by passing the lysate 20 times through a 20-gauge needle. After centrifugation (8000 rpm, 10 min, 4 °C), the supernatant was used for protein determination and further analysis. For total extracts, 500 μl of cold radioimmunoprecipitation buffer (RIPA: 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.5 mm DTT, 1000 units benzonase and supplemented with a pre-made protease inhibitor mixture (see above)) was added to 3 × 106 cells. Samples were vortexed and sonicated for 30 s (50% power, 3 cycles) with an ultrasonic homogenizer HD2070 (Bandelin). After 30 min of incubation, samples were centrifuged (14,000 rpm, 10 min, 4 °C), and supernatant was used for protein determination and further analysis. All protein concentrations were determined using the Bradford technique and BSA as calibration standard as described previously (52).
Western Blotting
20 μg of protein was incubated in 2× Laemmli buffer (Sigma) at 95 °C for 10 min and separated by a Mini-PROTEAN® TGX (Bio-Rad) precasted gel (4–20% acrylamide). Proteins were then transferred to a nitrocellulose membrane and blocked with 2% dry milk (in PBS) or in the case of protein modifications with a Pierce® protein-free blocking buffer (Thermo Fisher Scientific) at room temperature for 1 h. Membranes were then incubated overnight at 4 °C with antibodies against GLO1 (1:1000 dilution; ab81461; rat; Abcam), GSH reductase (1:1000 dilution; ab16801; rabbit; Abcam), aldo-keto reductase 1b1 (1:1000 dilution; ab175394; rabbit; Abcam), and β-actin (1:2500 dilution; 4967S; rabbit; Cell Signaling Technology) in 2% dry milk containing PBS and 0.05% Tween 20 (PBS-T). After three washing steps (5 min each) with PBS-T, membranes were incubated with horseradish-linked goat anti-rat (1:2000 dilution; 7077S; Cell Signaling Technology) or goat anti-rabbit (1:2000 dilution; 7077S; Cell Signaling Technology) antibody for 1 h at room temperature. Proteins were visualized on X-ray films using ECL detection reagents (GE Healthcare) with varying exposure times (0.1–2 min). Protein expression was determined using a densitometry analyzing software QuantityOne (Bio-Rad) and normalized to β-actin expression.
Methylglyoxal Toxicity Assays
Viability assays were performed with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT method) as described previously with some minor changes (53). Cells were seeded into 96-well plates and grown to 80% confluency under basal growth conditions. Cells were then customized to DMEM with only 0.1% FCS for 24 h and afterward exposed to increasing concentrations of MG (10–500 μm) for 48 h. For viability determination, 50 μl of MTT solution (2 mg/ml in PBS) was added to the medium, and cells were incubated at 33 °C for 2 h. Reduced MTT was then solubilized in 200 μl of DMSO, and absorbance was measured at 590 nm (reference at 690 nm) using a FLUOstar OMEGA multiplate reader (BMG Labtech). Viability was calculated as a percentage of controls (untreated cells). Data were fitted by nonlinear regression using the GraphPad PRISM 6 software (GraphPad Software Inc.), and values of LD50 were determined.
Determination of Intracellular GSH and GSSG
Cells were exposed to 0.2 mm HTA for 10, 20, 30, 60, 90, and 120 min. HTA was prepared according the Kd equations calculated previously (41). Quantitative analysis of glutathione and glutathione disulfide were performed using an enzymatic recycling method as described previously (54).
Detection of S-Nitrosylation and Production of S-Nitrosylated AKR1b3
S-Nitrosylation was detected by the iodoTMT switch technique, a modification of the biotin switch technique described elsewhere (55) using a PierceTM S-nitrosylation Western blotting kit (Thermo Fisher Scientific), according to the manufacturer's instructions. For the nitrosylation of recombinant AKR1b3, streptozotocin was used as an NO donor (56). To 1 μg of recombinant protein in sodium phosphate buffer (0.1 m (pH 7.2)), 0.1 mmol of streptozotocin was added and incubated for 30 min at room temperature. S-Nitrosylated recombinant protein was washed and purified using a spin filter column with a 3-kDa cutoff (Millipore). Nitrosylation of recombinant AKR1b3 was confirmed by Western blotting.
Enzymatic Activity Assays
Activity of GLO1 was determined spectrophotometrically as described previously (57). Briefly, the method monitors the initial rate of change in absorbance at 235 nm caused by the formation of S-d-lactoylglutathione through catalysis of GLO1. The assay mixture contained 2 mm MG and 2 mm GSH in sodium phosphate buffer (50 mm (pH 6.6), 37 °C) and was incubated for 15 min to guarantee the complete formation of HTA. After the addition of the cytosolic protein fraction (1 μg/μl), the change in absorbance was monitored for 15 min. The activity of GLO1 is described in units, where 1 unit is the amount of GLO1 that catalyzes the formation of 1 μmol of S-d-lactoylglutathione/min.
The kinetic activity assay for AKR in cytosolic fractions was performed as described previously with minor changes (58). Briefly, activity was determined in 0.1 m sodium phosphate buffer (pH 7.2) containing MG or HTA (0.1–2 mm) and 0.1 mm NADPH at 25 °C. The reaction was monitored by following the loss of NADPH at 340 nm. ALDH activity was assayed at 25 °C in 75 mm Tris-HCl (pH 9.5) containing 10 mm dl-2-amino-1-propanol, 0.5 mm NADP, and 0.1–2 mm MG by measuring the rate of NADP formation at 340 nm (12). Activity for GSH reductase was determined as described previously (59). For all kinetic analyses, the data were fitted by nonlinear regression using the GraphPad PRISM 6 software (GraphPad Software Inc.), and Km and Vmax values were calculated. Activity for NOS was determined using a commercially available kit (BioVision). In this assay, NO generated by NOS reacts with a fluorescent probe to generate a signal at excitation/emission = 360/450 nm, which is proportional to NOS activity.
Conversion of Labeled d4-MG by Wild-type and GLO1−/− Schwann Cells
Product analysis was achieved by exposing wild-type and GLO1−/− Schwann cells with 150 μm d4-MG (in DMEM with 0.1% FCS). Cells were then treated with 1 ml of ice-cold PBS and harvested using a cell scraper (Gibco). After the addition of 100 μl of trichloroacetic acid (20% w/v in 0.9% NaCl), lysates were vortexed and sonicated for 30 s (50% power, 3 cycles). After a short spin down (14,000 rpm, 5 min, 4 °C), supernatant was stored at −20 °C or used directly for further LC-MS/MS analysis.
Determination of Metabolites via LC-MS/MS
Hydroxyacetone, lactaldehyde, and lactate were determined using O-(2,3, 4,5,6-pentafluorophenyl) methyl hydroxylamine (PFBHA) as a derivatizing agent as described previously, with some changes (60). Supernatants (900 μl) from the previous method description were used for the derivatizing procedure (addition of 50 μl of PFBHA (5 mm) for 2 h at room temperature). After the derivatizing procedure, 10 μl of sulfuric acid was added to bind excessive PFBHA. Mixture was then mixed with 600 μl of n-hexane, which was isolated and evaporated using a vacuum concentrator (Savant SpeedVacTM SC100) at room temperature. Derivatized compounds were resolved in 100 μl of a H2O/acetonitrile mixture (50:50) and used for LC-MS/MS analysis.
An Acquity UPLC class 1 liquid chromatography system (Waters®) was used, and analytes were separated by reverse phase LC on a Waters® Acquity BEH C18 column (1.7 μm, 2.1 × 50 mm) at a flow rate of 0.3 ml/min and a column temperature of 40 °C. Gradient elution was performed using solvent A (99.9% water + 0.01% formic acid) and solvent B (99.9% acetonitrile + 0.01% formic acid). Gradient elution was performed with a total run time of 1 h to guarantee a separation of components with the same mass. The gradient started with 90% solvent A and was increased to 90% solvent B over 50 min, and then held at 90% solvent B for 5 min. The detection was carried out on a XEVO TQ-S tandem quadrupole mass spectrometer (Waters®) using positive electrospray ionization mode. Analyte detection was performed using multiple reaction monitoring (MRM). Source parameters were set as follows: capillary voltage 3.8 kV; desolvation temperature 300 °C; desolvation gas flow 850 liters/h; source temperature 150 °C; cone gas flow 250 liters/h; collision gas flow 0.15 ml/min; nebulizer gas flow 5 bar. Identification was based upon retention times, two MRM transitions for each compound (qualifier and quantifier) and the final comparison of standards with a defined concentration. MRM transitions used and retention times were as follows: hydroxyacetone (m/z 270.1 > 252.1 and m/z 270.1 > 181.1) (retention time 23.88 min); d4-hydroxyacetone (m/z 274.1 > 256.1 and m/z 274.1 > 181.1) (retention time 23.86 min); dl-lactate (m/z 284.2 > 266.1 and m/z 284.2 > 181.1) (retention time 18.42 min); d4-dl-lactate (m/z 288.2 > 270.1 and m/z 288.2 > 181.1) (retention time 18.43 min); lactaldehyde (m/z 270.1 > 252.1 and m/z 270.1 > 181.1) (retention time 25.19 min); d4-lactaldehyde (m/z 274.1 > 256.1 and m/z 274.1 > 181.1) (retention time 25.17 min). Identification of products after the conversion of d4-MG was based on the comparison of retention times and the expected mass transitions of non-deuterated calibrators with a defined concentration. Acquisition and quantification were completed with MassLynx 4.1 and TargetLynx 2.7 (Waters®). The determination of MG and MG-derived AGEs by stable isotopic dilution analysis via LC-MS/MS was described previously (61, 62).
Determination of Reactive Oxygen, Superoxide, and Nitric Oxide Species
Determination was based upon analysis via flow cytometry/FACS. All incubation and washing steps of living cells were done in Krebs-Ringer HEPES buffer (KRH), including 136 mm NaCl, 4.7 mm KCl, 1.25 mm CaCl2, 1.25 mm MgSO4, 10 mm HEPES, 0.1% fatty acid-free BSA (pH 7.4). Treated and untreated cells were stained with Hoechst 33258 NucBlue® (Thermo Fisher Scientific) for the detection of viable cells. Determination of superoxide production and reactive oxygen species was achieved incubating Schwann cells with MitoSOXTM Red (10 μm in KRH buffer) and CM-H2DCFDA (5 μm in KRH buffer) for 30 min under reduced light conditions. For the determination of nitric oxide species we used 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM; 5 μm in KRH buffer) (all reagents from Thermo Fisher Scientific). After two washing steps, the cells were trypsinized and resuspended in 1 ml of FACS-Buffer (10% FCS, 1 mm EDTA in PBS). Analysis of fluorophores was performed using a LSRII flow cytometer (BD Biosciences) by gating initial cell population via forward scatter against side scatter signals and detecting viable cells (Hoechst positive) by violet laser (excitation, 405 nm; filter, 450/40 nm). Hoechst-positive cells were then analyzed for MitoSOXTM Red by blue laser (excitation, 488 nm; filter, 575/26 nm), CM-H2DCFDA, and DAF-FM also by blue laser (excitation, 488 nm; filter, 530/30 nm).
Statistical Analysis
All data are expressed as mean values ± S.E. and were analyzed for significance using two-tailed unpaired t test with Welch's correction. The comparison of more than one group was achieved using an ordinary one- or two-way ANOVA followed by comparing all groups using Tukey's (one-way ANOVA) or Sidak's (two-way ANOVA) multiple comparison test. Differences were considered significant at p < 0.05.
Author Contributions
J. M. performed and analyzed most of the experiments and wrote the manuscript. T. F. assisted in experimental preparation and in analysis or interpretation of data. D. S. and M. F. assisted in research design and performed initial experiments for the establishment of GLO1 knock-outs. V. E. assisted in performing experiments with flow cytometry/fluorescence-activated cell sorting (FACS). P. N. and S. H. conceived the idea of this project and assisted in writing the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Barbro Beijer and Prof. Walter Mier from the Department of Nuclear Medicine, University of Heidelberg, for the synthesis of d4-MG and Dr. Karel Klika from the German Cancer Research Center (DKFZ) for the NMR analysis. We also thank members of the Nawroth/Fleming laboratory for helpful comments and discussions.
This work was supported in whole or part by Deutsche Forschungsgemeinschaft Grants SFB1118 and GRK 1874-DIAMICOM and The DietmarHopp Foundation. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Materials and Methods.
- AGE
- advanced glycation end product
- 1400 W dihydrochloride
- N-([3-(aminomethyl)phenyl]methyl)ethanimidamide dihydrochloride
- 2-AAPA
- R,R′-2-acetylamino-3-[4-(2-acetylamino-2-carboxyethyl-sulfanylthiocarbonylamino)phenylthio-carbamoylsulfanyl]propionic acid hydrate
- AKR
- aldo-keto reductase
- ALDH
- aldehyde dehydrogenase
- HTA
- hemithioacetal
- KRH
- Krebs Ringer HEPES buffer
- MG
- methylglyoxal
- MG-H1
- methylglyoxal-derived hydroimidazolone 1
- PFBHA
- O-(2,3,4,5,6-pentafluorophenyl)methyl hydroxylamine
- ANOVA
- analysis of variance
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- DAF-FM
- 7′-difluorofluorescein diacetate
- iNOS
- inducible NOS
- qPCR
- quantitative PCR
- MRM
- multiple reaction monitoring
- MG-H1
- methylglyoxal-derived hydroimidazolone
- iodoTMT
- iodoacetyl tandem mass tag
- CM-H2DCFDA
- chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate.
References
- 1. Goh S.-Y., and Cooper M. E. (2008) The role of advanced glycation end products in progression and complications of diabetes. J. Clin. Endocrinol. Metab. 93, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 2. Singh R., Barden A., Mori T., and Beilin L. (2001) Advanced glycation end-products: a review. Diabetologia 44, 129–146 [DOI] [PubMed] [Google Scholar]
- 3. Thornalley P. J., Langborg A., and Minhas H. S. (1999) Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem. J. 344, 109–116 [PMC free article] [PubMed] [Google Scholar]
- 4. Phillips S. A., Mirrlees D., and Thornalley P. J. (1993) Modification of the glyoxalase system in streptozotocin-induced diabetic rats: effect of the aldose reductase inhibitor Statil. Biochem. Pharmacol. 46, 805–811 [DOI] [PubMed] [Google Scholar]
- 5. Han Y., Randell E., Vasdev S., Gill V., Gadag V., Newhook L. A., Grant M., and Hagerty D. (2007) Plasma methylglyoxal and glyoxal are elevated and related to early membrane alteration in young, complication-free patients with type 1 diabetes. Mol. Cell. Biochem. 305, 123–131 [DOI] [PubMed] [Google Scholar]
- 6. McLellan A. C., Thornalley P. J., Benn J., and Sonksen P. H. (1994) Glyoxalase system in clinical diabetes mellitus and correlation with diabetic complications. Clin. Sci. 87, 21–29 [DOI] [PubMed] [Google Scholar]
- 7. Allaman I., Bélanger M., and Magistretti P. J. (2015) Methylglyoxal, the dark side of glycolysis. Front. Neurosci. 9, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thornalley P. J. (1990) The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 269, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thornalley P. J. (2003) Glyoxalase I—Structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 31, 1343–1348 [DOI] [PubMed] [Google Scholar]
- 10. Brouwers O., Niessen P. M., Ferreira I., Miyata T., Scheffer P. G., Teerlink T., Schrauwen P., Brownlee M., Stehouwer C. D., and Schalkwijk C. G. (2011) Overexpression of glyoxalase-I reduces hyperglycemia-induced levels of advanced glycation end products and oxidative stress in diabetic rats. J. Biol. Chem. 286, 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bélanger M., Yang J., Petit J.-M., Laroche T., Magistretti P. J., and Allaman I. (2011) Role of the glyoxalase system in astrocyte-mediated neuroprotection. J. Neurosci. 31, 18338–18352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skapare E., Konrade I., Liepinsh E., Strele I., Makrecka M., Bierhaus A., Lejnieks A., Pirags V., and Dambrova M. (2013) Association of reduced glyoxalase 1 activity and painful peripheral diabetic neuropathy in type 1 and 2 diabetes mellitus patients. J. Diabetes Complicat. 27, 262–267 [DOI] [PubMed] [Google Scholar]
- 13. Jack M. M., Ryals J. M., and Wright D. E. (2011) Characterisation of glyoxalase 1 in a streptozotocin-induced mouse model of diabetes with painful and insensate neuropathy. Diabetologia 54, 2174–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peters A. S., Lercher M., Fleming T. H., Nawroth P. P., Bischoff M. S., Dihlmann S., Böckler D., and Hakimi M. (2016) Reduced glyoxalase 1 activity in carotid artery plaques of nondiabetic patients with increased hemoglobin A1c level. J. Vasc. Surg. 64, 990–994 [DOI] [PubMed] [Google Scholar]
- 15. Bierhaus A., Fleming T., Stoyanov S., Leffler A., Babes A., Neacsu C., Sauer S. K., Eberhardt M., Schnölzer M., Lasitschka F., Lasischka F, Neuhuber W. L., Kick T. I., Konrade I., Elbert R., et al. (2012) Methylglyoxal modification of Na(v) 1.8 facilitates nociceptive neuron firing and causes hypoalgesia in diabetic neuropathy. Nat. Med. 18, 926–933 [DOI] [PubMed] [Google Scholar]
- 16. Thornalley P. J., Bata S., Ahmed N., Karachalias N., Agalou S., Babaei-Jadidi R., and Dawnay A. (2003) Quantitative screening of advanced glycation end products in cellular and extracellular proteins by tandem mass spectrometry. Biochem. J. 375, 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freeman O. J., Unwin R. D., Dowsey A. W., Begley P., Ali S., Hollywood K. A., Rustogi N., Petersen R. S., Dunn W. B., Cooper G. J., and Gardiner N. J. (2016) Metabolic dysfunction is restricted to the sciatic nerve in experimental diabetic neuropathy. Diabetes 65, 228–238 [DOI] [PubMed] [Google Scholar]
- 18. Vander Jagt D. L., and Hunsaker L. A. (2003) Methylglyoxal metabolism and diabetic complications: role of aldose reductase, glyoxalase-I, betaine aldehyde dehydrogenase, and 2-oxoaldehyde dehydrogenase. Chem. Biol. Interact. 143, 341–351 [DOI] [PubMed] [Google Scholar]
- 19. Vander Jagt D. L., Robinson B., Taylor K. K., and Hunsaker L. A. (1992) Reduction of trioses by NADPH-dependent aldo-keto reductase. J. Biol. Chem. 267, 4364–4369 [PubMed] [Google Scholar]
- 20. Baba S. P., Barski O. A., Ahmed Y., O'Toole T. E., Conklin D. J., Bhatnagar A., and Srivastava S. (2009) Reductive metabolism of AGE precursors: a metabolic route for preventing AGE accumulation in cardiovascular tissue. Diabetes 58, 2486–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Izaguirre G., Kikonyogo A., and Pietruszko R. (1998) Methylglyoxal as substrate and inhibitor of human aldehyde dehydrogenase: comparison of kinetic properties among the three isozymes. Comp. Biochem. Physiol. B Biochem Mol. Biol. 119, 747–754 [DOI] [PubMed] [Google Scholar]
- 22. Lee J. Y., Song J., Kwon K., Jang S., Kim C., Baek K., Kim J., and Park C. (2012) Human DJ-1 and its homologs are novel glyoxalases. Hum. Mol. Genet. 21, 3215–3225 [DOI] [PubMed] [Google Scholar]
- 23. Schemmel K. E., Padiyara R. S., and D'Souza J. J. (2010) Aldose reductase inhibitors in the treatment of diabetic peripheral neuropathy: a review. J. Diabetes Complicat. 24, 354–360 [DOI] [PubMed] [Google Scholar]
- 24. Ramirez M. A., and Borja N. L. (2008) Epalrestat: an aldose reductase inhibitor for the treatment of diabetic neuropathy. Pharmacotherapy 28, 646–655 [DOI] [PubMed] [Google Scholar]
- 25. Mizisin A. P. (2014) Mechanisms of diabetic neuropathy. Handb. Clin. Neurol. 126, 401–428 [DOI] [PubMed] [Google Scholar]
- 26. Viader A., Sasaki Y., Kim S., Strickland A., Workman C. S., Yang K., Gross R. W., and Milbrandt J. (2013) Aberrant Schwann cell lipid metabolism linked to mitochondrial deficits leads to axon degeneration and neuropathy. Neuron 77, 886–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hidmark A., Fleming T., Vittas S., Mendler M., Deshpande D., Groener J. B., Müller B. P., Reeh P. W., Sauer S. K., Pham M., Muckenthaler M. U., Bendszus M., and Nawroth P. P. (2014) A new paradigm to understand and treat diabetic neuropathy. Exp. Clin. Endocrinol. Diabetes 122, 201–207 [DOI] [PubMed] [Google Scholar]
- 28. Rabbani N., and Thornalley P. J. (2014) The critical role of methylglyoxal and glyoxalase 1 in diabetic nephropathy. Diabetes 63, 50–52 [DOI] [PubMed] [Google Scholar]
- 29. Giacco F., Du X., D'Agati V. D., Milne R., Sui G., Geoffrion M., and Brownlee M. (2014) Knockdown of glyoxalase 1 mimics diabetic nephropathy in nondiabetic mice. Diabetes 63, 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shinohara M., Thornalley P. J., Giardino I., Beisswenger P., Thorpe S. R., Onorato J., and Brownlee M. (1998) Overexpression of glyoxalase-I in bovine endothelial cells inhibits intracellular advanced glycation end product formation and prevents hyperglycemia-induced increases in macromolecular endocytosis. J. Clin. Invest. 101, 1142–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xue M., Rabbani N., Momiji H., Imbasi P., Anwar M. M., Kitteringham N., Park B. K., Souma T., Moriguchi T., Yamamoto M., and Thornalley P. J. (2012) Transcriptional control of glyoxalase 1 by Nrf2 provides a stress-responsive defence against dicarbonyl glycation. Biochem. J. 443, 213–222 [DOI] [PubMed] [Google Scholar]
- 32. Xue M., Weickert M. O., Qureshi S., Kandala N.-B., Anwar A., Waldron M., Shafie A., Messenger D., Fowler M., Jenkins G., Rabbani N., and Thornalley P. J. (2016) Improved glycemic control and vascular function in overweight and obese subjects by glyoxalase 1 inducer formulation. Diabetes 65, 2282–2294 [DOI] [PubMed] [Google Scholar]
- 33. Su Y., Qadri S. M., Wu L., and Liu L. (2013) Methylglyoxal modulates endothelial nitric oxide synthase-associated functions in EA.hy926 endothelial cells. Cardiovasc. Diabetol. 12, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turkseven S., Ertuna E., Yetik-Anacak G., and Yasa M. (2014) Methylglyoxal causes endothelial dysfunction: the role of endothelial nitric oxide synthase and AMP-activated protein kinase α. J. Basic Clin. Physiol. Pharmacol. 25, 109–115 [DOI] [PubMed] [Google Scholar]
- 35. Chang T., Wang R., and Wu L. (2005) Methylglyoxal-induced nitric oxide and peroxynitrite production in vascular smooth muscle cells. Free Radic. Biol. Med. 38, 286–293 [DOI] [PubMed] [Google Scholar]
- 36. Kaiserova K., Tang X.-L., Srivastava S., and Bhatnagar A. (2008) Role of nitric oxide in regulating aldose reductase activation in the ischemic heart. J. Biol. Chem. 283, 9101–9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baba S. P., Wetzelberger K., Hoetker J. D., and Bhatnagar A. (2009) Posttranslational glutathiolation of aldose reductase (AKR1B1): a possible mechanism of protein recovery from S-nitrosylation. Chem. Biol. Interact. 178, 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vander Jagt D. L., Hassebrook R. K., Hunsaker L. A., Brown W. M., and Royer R. E. (2001) Metabolism of the 2-oxoaldehyde methylglyoxal by aldose reductase and by glyoxalase-I: roles for glutathione in both enzymes and implications for diabetic complications. Chem. Biol. Interact. 130, 549–562 [DOI] [PubMed] [Google Scholar]
- 39. Rae C., Berners-Price S. J., Bulliman B. T., and Kuchel P. W. (1990) Kinetic analysis of the human erythrocyte glyoxalase system using 1H NMR and a computer model. Eur. J. Biochem. 193, 83–90 [DOI] [PubMed] [Google Scholar]
- 40. Vander Jagt D. L. (1989) in Glutathione: Chemical, Biochemical, and Medical Aspects (Dolphin D., Paulson R., and Avramovic O., eds) Part A, pp. 598–641, Wiley and Sons, New York [Google Scholar]
- 41. Thornalley P. J. (1993) The glyoxalase system in health and disease. Mol. Aspects Med. 14, 287–371 [DOI] [PubMed] [Google Scholar]
- 42. Barski O. A., Tipparaju S. M., and Bhatnagar A. (2008) The Aldo-Keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab. Rev. 40, 553–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brownlee M. (2001) Biochemistry and molecular cell biology of diabetic complications. Nature 414, 813–820 [DOI] [PubMed] [Google Scholar]
- 44. Kern T. S., and Engerman R. L. (1982) Immunohistochemical distribution of aldose reductase. Histochem. J. 14, 507–515 [DOI] [PubMed] [Google Scholar]
- 45. Sango K., Kato K., Tsukamoto M., Niimi N., Utsunomiya K., and Watabe K. (2014) Physiological and pathological roles of aldose reductase in Schwann cells. J. Mol. Genet. Med. S1, 012 [Google Scholar]
- 46. Hu X., Li S., Yang G., Liu H., Boden G., and Li L. (2014) Efficacy and safety of aldose reductase inhibitor for the treatment of diabetic cardiovascular autonomic neuropathy: systematic review and meta-analysis. PLoS ONE 9, e87096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clelland J. D., and Thornalley P. J. (1990) Synthesis of 14C-labelled methylglyoxal and S-d-lactoylglutathione. J. Label. Comp. Radiopharm. 28, 1455–1464 [Google Scholar]
- 48. Wang T., Kartika R., and Spiegel D. A. (2012) Exploring post-translational arginine modification using chemically synthesized methylglyoxal hydroimidazolones. J. Am. Chem. Soc. 134, 8958–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ahmed N., Dobler D., Dean M., and Thornalley P. J. (2005) Peptide mapping identifies hotspot site of modification in human serum albumin by methylglyoxal involved in ligand binding and esterase activity. J. Biol. Chem. 280, 5724–5732 [DOI] [PubMed] [Google Scholar]
- 50. Goedhart J., von Stetten D., Noirclerc-Savoye M., Lelimousin M., Joosen L., Hink M. A., van Weeren L., Gadella T. W. Jr., and Royant A. (2012) Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 3, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 52. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 53. Denizot F., and Lang R. (1986) Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 89, 271–277 [DOI] [PubMed] [Google Scholar]
- 54. Rahman I., Kode A., and Biswas S. K. (2006) Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 1, 3159–3165 [DOI] [PubMed] [Google Scholar]
- 55. Qu Z., Meng F., Bomgarden R. D., Viner R. I., Li J., Rogers J. C., Cheng J., Greenlief C. M., Cui J., Lubahn D. B., Sun G. Y., and Gu Z. (2014) Proteomic quantification and site-mapping of S-nitrosylated proteins using isobaric iodoTMT reagents. J. Proteome Res. 13, 3200–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kwon N. S., Lee S. H., Choi C. S., Kho T., and Lee H. S. (1994) Nitric oxide generation from streptozotocin. FASEB J. 8, 529–533 [DOI] [PubMed] [Google Scholar]
- 57. McLellan A. C., and Thornalley P. J. (1989) Glyoxalase activity in human red blood cells fractioned by age. Mech. Ageing Dev. 48, 63–71 [DOI] [PubMed] [Google Scholar]
- 58. Srivastava S., Watowich S. J., Petrash J. M., Srivastava S. K., and Bhatnagar A. (1999) Structural and kinetic determinants of aldehyde reduction by aldose reductase. Biochemistry 38, 42–54 [DOI] [PubMed] [Google Scholar]
- 59. Carlberg I., and Mannervik B. (1985) Glutathione reductase. Methods Enzymol. 113, 484–490 [DOI] [PubMed] [Google Scholar]
- 60. Beilin E., Baker L. J., Culbert P., and Toltl N. P. (2008) Quantitation of acetol in common pharmaceutical excipients using LC-MS. J. Pharm. Biomed. Anal. 46, 316–321 [DOI] [PubMed] [Google Scholar]
- 61. Rabbani N., and Thornalley P. J. (2014) Measurement of methylglyoxal by stable isotopic dilution analysis LC-MS/MS with corroborative prediction in physiological samples. Nat. Protoc. 9, 1969–1979 [DOI] [PubMed] [Google Scholar]
- 62. Thornalley P. J., and Rabbani N. (2014) Detection of oxidized and glycated proteins in clinical samples using mass spectrometry—a user's perspective. Biochim. Biophys. Acta 1840, 818–829 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.