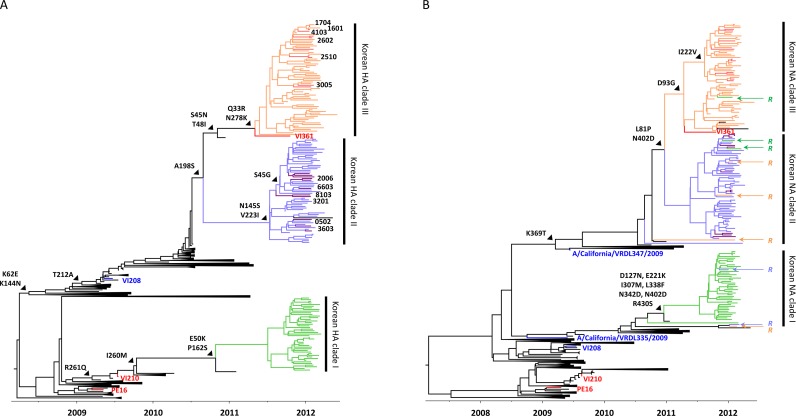
Fig 2. Phylogenetic relationships of the HA and NA genes of Korean H3N2 strains circulating in the 2011/12 season.
The evolutionary relationships of the HA and NA genes of Korean H3N2 strains were reconstructed using the KISED (n = 154) and KUMC (n = 21) sequences. The sequences downloaded from the NCBI (n = 451) and GISAID (n = 8) databases were used as references. The three Korean HA and NA clades are indicated with different colors (clade I, light green; clade II, violet; and clade III, orange), and the KUMC sequences in the clades II and III were highlighted with purple and light red colors, respectively. The vaccine virus tips (PE16, VI210, and VI361) and the other clade-defining sequences (VI208, A/California/VRDL335/2009, and A/California/VRDL347/2009) were colored with red and blue, respectively. The KUMC viruses used for the HI assay were indicated with viral isolate numbers (please see Table 1). In the NA tree, the sequences of reassortment candidate strains between the HA and NA genes were indicated with a capital letter ‘R’ and the colors of their corresponding HA clades. Trunk lineage amino acid mutations were also indicated.