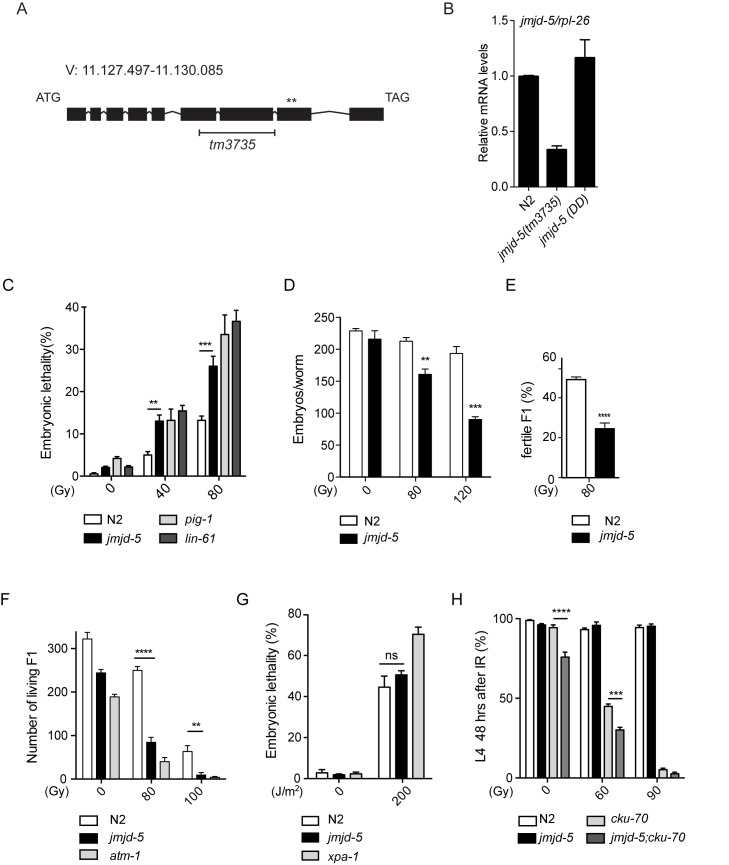
Fig 1. jmjd-5 mutant animals are hypersensitive to IR.
(A) Genomic structure of the jmjd-5 gene. Dark boxes represent coding sequences and lines represent introns. The black H-shaped bar specifies the deletion in the tm3735 allele and the asterisks indicate the position of the two amino acids (H484 and D486) located in the JmjC domain, mutagenized in jmjd-5(DD). (B) Quantification of jmjd-5 mRNA levels by qRT-PCR in the indicated strains. rpl-26 is used as internal control. (C) Percent of embryonic lethality in the indicated strains observed after irradiation of young adult worms (24 h post L4) with 0, 40, or 80 Gy of IR. (D) Brood size in N2 and jmjd-5(tm3735) after IR treatment of young adults with 0, 80, or 120 Gy. Deposited embryos are counted from the time of irradiation. (E) Percentage of fertile animals in N2 and jmjd-5(tm3735) (evaluated by the presence of embryos in the uterus) among the living F1 progeny of irradiated mothers. (F) Number of living F1 animals in the indicated strains after irradiation of L1 larvae with 0, 80, or 100 Gy of IR. L1 were irradiated and the total number of living F1 offspring that passed the L1 stage is reported. (G) Percent of embryonic lethality in the indicated strains observed after treatment of young adult worms with 0 or 200 J/m2 of UV. (H) Percent of irradiated L1 of the indicated strains that developed to the L4 stage in 48 hours. In C-H, pig-1(gm344), lin-61(n3809), atm-1(gk186), cku-70(tm1524) and xpa-1(ok698) are used as positive controls. In B-H, data are represented as means ± sem from at least 3 biological independent experiments. ****p< 0.0001, ***p<0.005, **p<0.01, n.s. = not significant, with two-tailed, unpaired t-test.