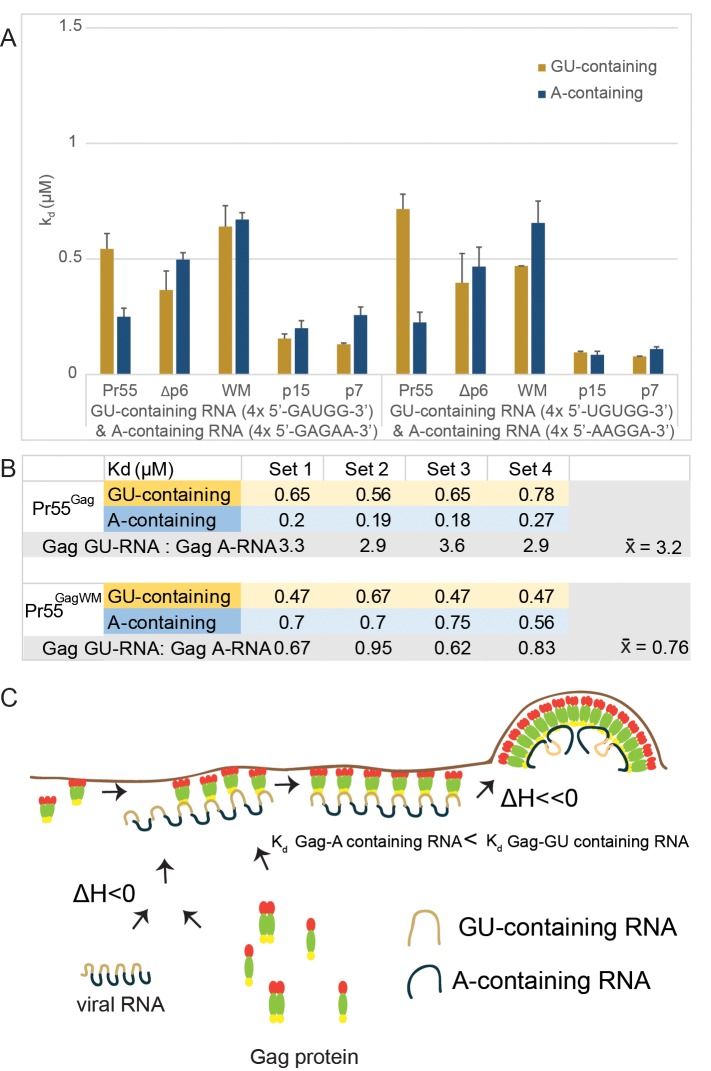
Fig 5. Oligomeric Gag has increased binding preference towards A-containing over GU-containing RNA sequences.
(A) Calculated binding affinities (Kd) from ITC binding curves fitted to a one-site interaction model are plotted for replicate interactions of first set of GU-containing and A-containing RNA (4x 5’-GUAGG-3’ and 4x 5’-GAGAA-3’, respectively) and second set of GU-containing and A-containing RNA (4x 5’-UGUGG-3’ and 4x 5’-AAGGA-3’, respectively) with Pr55Gag, Pr50GagΔp6, Pr55GagWM, p15NC-SP2-p6 and p7NC. (B) Calculated binding affinities (Kd) for Pr55Gag and Pr55GagWM for replicate sets of interactions were tabulated and the fold difference between GU-containing and A-containing RNA binding affinities for each set were calculated. The average fold difference across the 4 sets is highlighted in bold. Sets 1 and 2 represent Kd values obtained from replicate Gag interactions with 4x 5’-GUAGG-3’ (GU-containing RNA) and 4x 5’-GAGAA-3’ (A-containing RNA). Sets 3 and 4 represent Kd values obtained from replicate Gag interactions with a second set of GU-containing and A-containing RNA (4x 5’-UGUGG-3’ and 4x 5’-AAGGA-3’, respectively). (C) Proposed model for Pr55Gag trafficking and viral RNA interaction during viral assembly. The schematic representation of GU-containing and A-containing RNA motifs aims to represent how Gag interacts with different RNA motifs during virus biogenesis. The distributions of these RNA motifs are not evenly spread-out across HIV RNA genome, and more precise distributions of these RNA can be seen in S3 Fig.