Fig. 5. Schematic model of signal integration pathways in phytochrome-linked enzymatic effectors and in-solution structure of IsPadC.
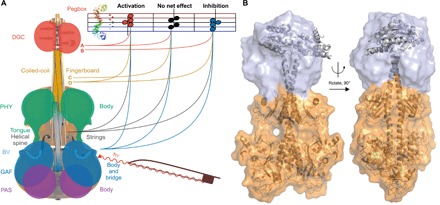
(A) The characteristic structure of IsPadC and regulatory properties of closely related homologs suggest a model of signal transduction corresponding to the violin model (28). Instead of a linear cascade of structural changes resulting in enzyme activation, the conformational dynamics of the whole system define the population of functionally relevant states, leading to either activation or inactivation with similar overall architectures (37). In the case of the phytochrome-violin, the pegbox corresponds to the effector domain, whose activity is tuned by the sensory module. Hitting the right chords on the enzymatic activity clef for stimulating GTP turnover is more complex than a specific actuation of the fingerboard, which would correspond to, for example, variation of its length (yellow lines and linkers C and D), and additionally depends on properties of the strings (gray), the shape of the violin body (blue), and the effector-pegbox (red; for example, two different DGC constructs A and B). The characteristic structural changes observed for various phytochrome structures (Fig. 4), which had also been interpreted as specific light-induced rearrangements (20, 24), reflect the structural plasticity of phytochrome dimerization. The latter, in turn, enables the modulation of the conformational dynamics of the overall system and thereby allows complex, evolutionary fine-tuning of the body of phytochrome-violin to optimize the output functionality as required by each system. The absence of characteristic structural changes, such as a defined rotation or a separation of the coiled-coil linker, enables the realization of systems featuring activation or inactivation within the same molecular architecture. (B) SAXS-based structural model for the IsPadC dimer in its dark-adapted Pr state. The surface represents the conformational space sampled by the GGDEF domains in the seven best structures according to the fit between the experimental and back-calculated SAXS data (compare fig. S10). Structures are aligned to the PAS-GAF-PHY domains.
