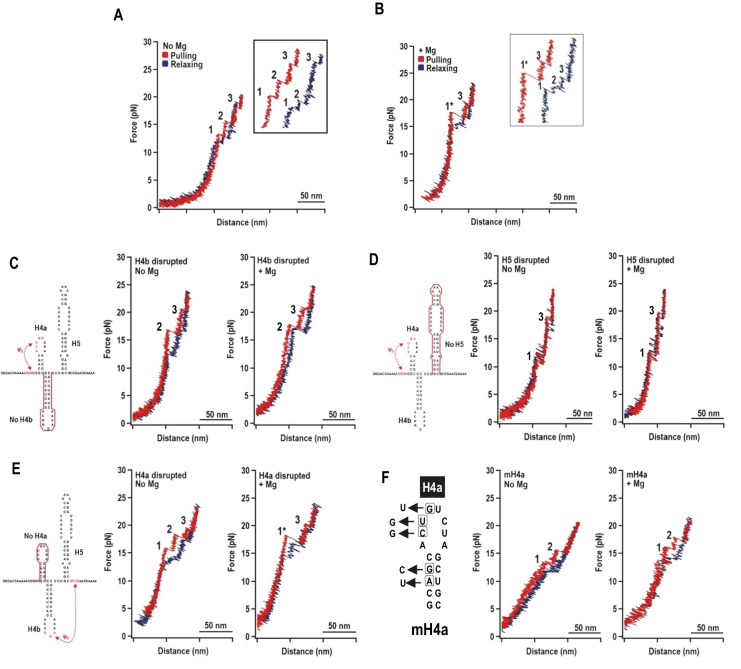
Figure 2. Representative force-extension curves (FEC) for the TSS using force ramping.
(A) FEC of the TSS in the absence of Mg2+ (N = 86). The red line represents the unfolding process and blue line represents the refolding process. The three intermediates are labeled. Insets show enlarged transitions. (B) FEC of TSS in Mg2+ (N = 64). The large and small rips in the unfolding process are labeled 1* and 3, respectively. (C, D, E) TSS was annealed with oligonucleotides complementary to H4b, H5, and H4a, respectively. FECs were conducted in the absence (middle) and presence (right) of Mg2+. The complimentary oligonucleotides are denoted with a red line. Note that the oligonucleotide complementary to H4a has no effect on the FEC. (F) FECs of TSS containing mutations in H4a (mH4a). All rips are numbered as for wt TSS.
Figure 2—figure supplement 1. Optical tweezer (OT) experimental set-up.

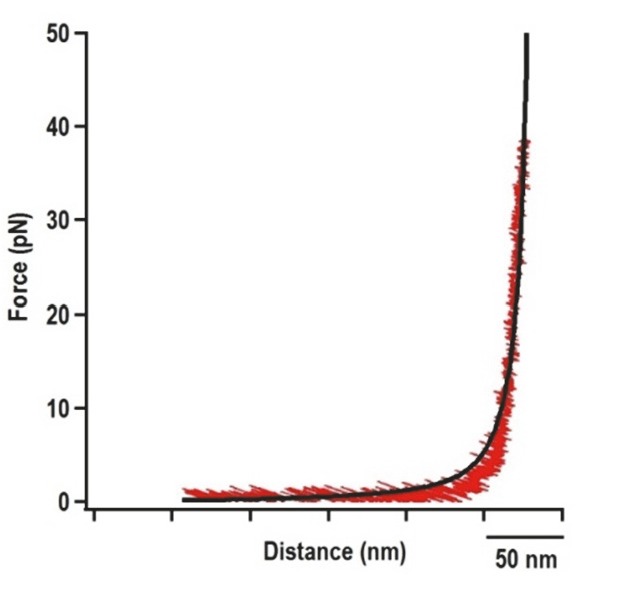
Figure 2—figure supplement 2. Force extension curve (FEC) of the DNA/RNA handles alone.