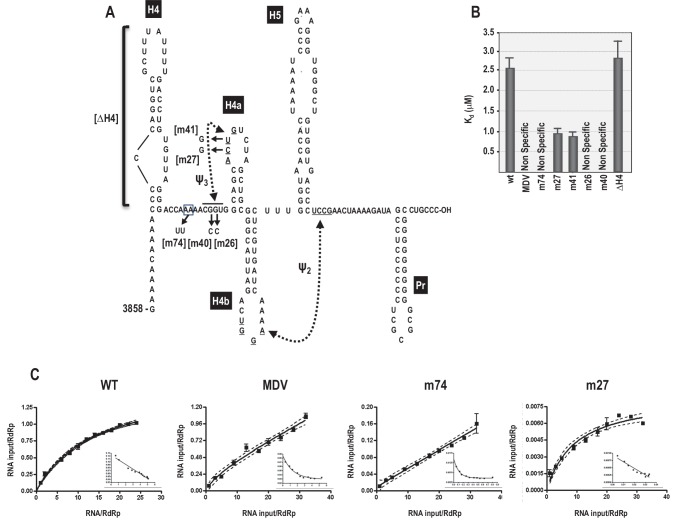
Figure 7. RdRp binding to mutant and wt 3’ fragments in vitro.
(A) Sequence and structure of the fragment used for RdRp binding. Locations and names of alterations are shown. ΔH4 is a complete deletion of hairpin H4. Numbering is from the 5’ end of the viral genomic RNA. (B) Five to 160 pmol of labeled wt and mutant fragments or control MDV RNA was combined with 11.4 pmol of purified, recombinant TCV RdRp, and bound RNA detected following filtration. MDV is a subviral RNA associated with Qβ bacteriophage and serves as a negative control for RdRp binding. (C) Selected saturation binding curves are shown together with Scatchard plots (insets). The similarity in non-linear Scatchard plots between MDV and m74 is interpreted as non-specific. For all assays, Kd were calculated from three independent experiments conducted in triplicate. Standard error bars are shown.