Figure 1. Conceptual diagram illustrating the various steps involved in estimating the mdNLR.
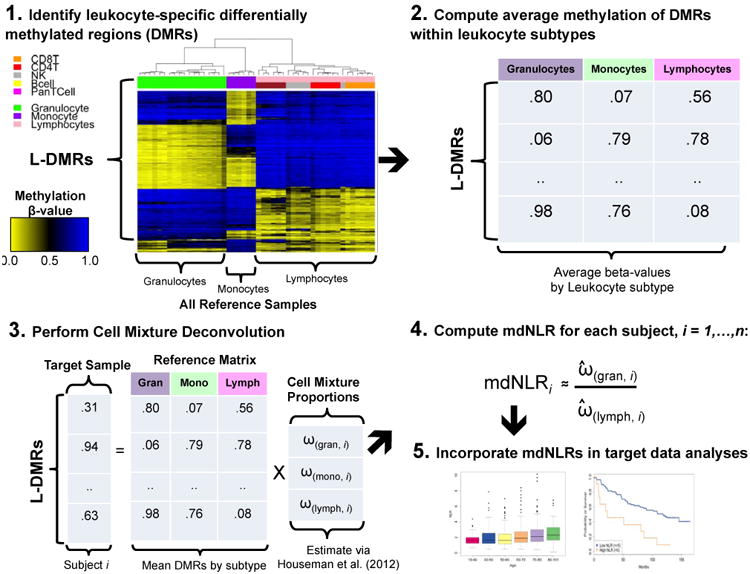
In Step 1, putative leukocyte differentially methylated regions (L-DMRs) are identified between monocytes, granulocytes, and lymphocytes. Step 2 involves computing the within cell type mean methylation beta-values for each of the putative L-DMRs identified in Step 1. In Step 3, the within cell type mean methylation beta-values are used in conjunction with Houseman et al., (23) to predict the proportion monocytes, granulocytes, and lymphocytes for a sample consisting of DNA methylation signatures profiled in whole blood. In Step 4, the mdNLR is calculated as the ratio of the predicted proportions of granulocytes and lymphocytes and finally, in Step 5 the estimated mdNLR is examined with respect to its association with cancer risk, outcomes, or other clinical variables of interest.
