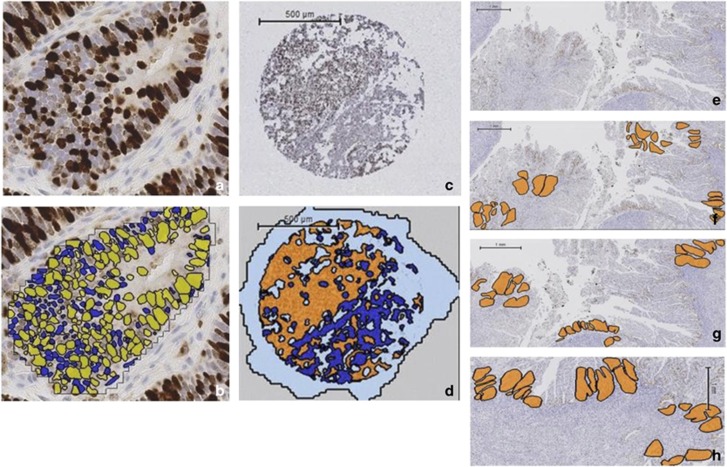
Figure 1.
Ki-67 immunohistochemistry and scoring using Definiens Developer on whole sections and tissue microarrays. The accuracy of the solution to correctly identify individual positively and negatively stained nuclei was manually checked by comparing individual endometrial cancer glands with and without the solution applied. (a) Photomicrograph of endometrial cancer gland with Ki-67 immunohistochemistry applied (b) digital scoring output (× 20 magnification). Positively stained nuclei are yellow, negatively stained nuclei are blue. (c) Representative tissue microarray core following Ki-67 immunohistochemistry. (d) Same tissue microarray core following the application of Definiens Developer solution, with endometrial cancer glands shown in orange and the surrounding stroma in dark blue. Areas of the slide without the presence of tissue are colored pale blue. (e) Whole section of tumor following Ki-67 immunohistochemistry. Compared with the tissue microarray, a significantly greater tumor area is present on a whole section and is a better representation of the heterogeneity in proliferation seen across endometrial cancers. (f) The same section of tumor with the five areas selected at random using Definiens Developer software highlighted in orange to determine the whole slide score. (g) Three areas of greatest proliferation identified to provide a hot spot score. (h) Three areas along the endometrial/myometrial interface to quantify the invasive edge score.