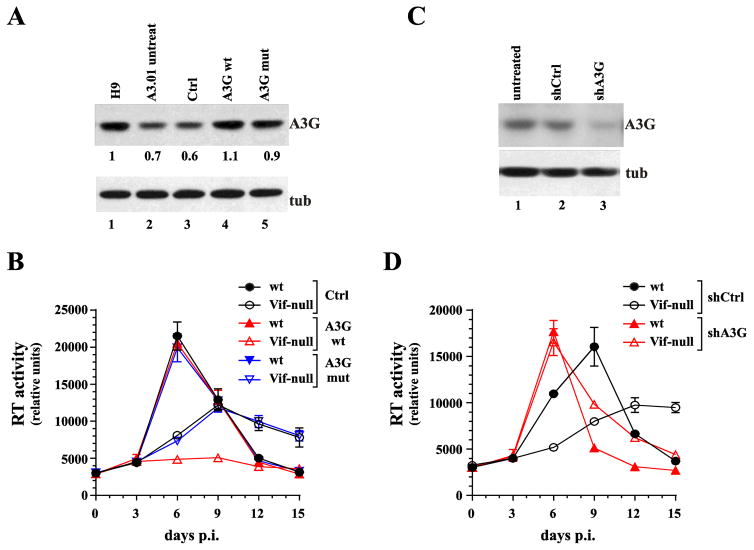
Fig. 3.
Increasing A3G levels in A3.01 cells induces a non-permissive phenotype whereas silencing of A3G produces a fully permissive phenotype. (A) A3.01 cells were transduced with lentiviral vectors encoding untagged wild type A3G (lane 4) or a deaminase-defective mutant (A3G C288S, C291A) (lane 5). Cells transduced with empty vector were analyzed in parallel (lane 3). Untreated H9 cells (lane 1) and A3.01 cells (lane 2) were analyzed in parallel. After transduction, cells were cultured for 48 h prior to puromycin selection. Puromycin-resistant A3.01 cells were amplified and A3G expression was determined by immunoblotting using untreated parental A3.01 cells and endogenous tubulin (tub) as reference. Protein gels were quantified by densitometry. (B) Transduced A3.01 cells were infected with wild type and Vif-null virus and replication profiles were determined by measuring supernatant reverse transcriptase levels over a 15-day period. Data are presented as mean +/− SD calculated from triplicate infections. (C) A3G was knocked down by transducing A3.01 cells with A3G shRNA encoding lentiviral vectors (lane 3). As a control, cells were treated with a vector encoding a non-silencing shRNA (lane 2). Puromycin-resistant cells were selected as described for panel A. Successful knockdown of A3G was verified by immunoblotting using an A3G specific polyclonal antibody. Parental A3.01 cells (untreated) served as specificity control (lane 1). The blot was reprobed with antibodies to α-tubulin to control for sample loading. (D) Puromycin-resistant knock-down cells and control cells were infected with wild type or NL4-3 Vif-null and replication profiles were determined as in panel B. Data are presented as mean +/− SD calculated from triplicate infections.