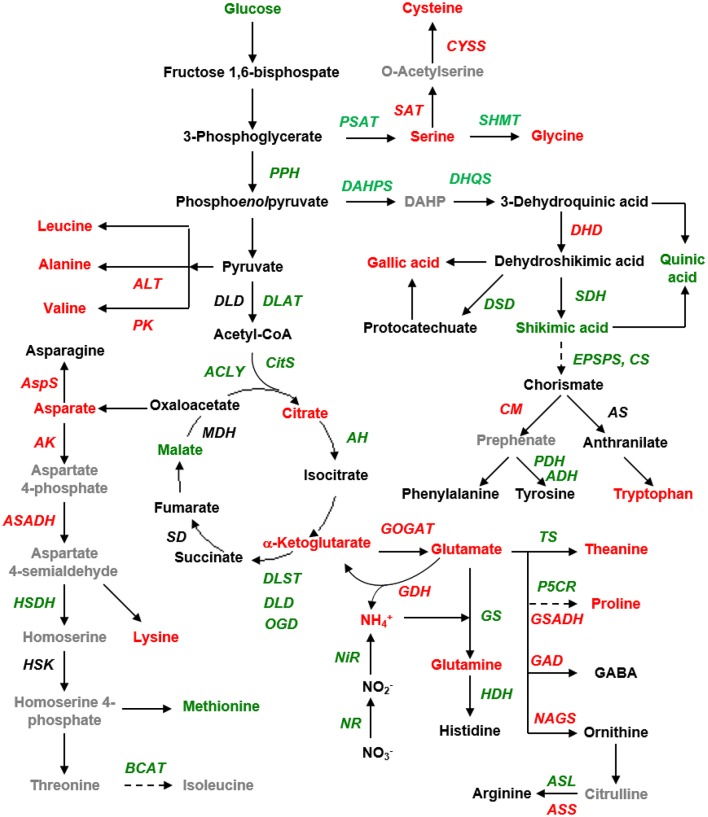
Figure 3.
Schematic presentation of the tricarboxylic acid cycle (TCA) pathway and amino acid biosynthesis, as affected by the chlorotic mutation. Red and green fonts indicate the up- and down-regulated genes and metabolites in the chlorotic leaves as compared with those in the non-chlorotic leaves. The gray font indicates the genes and metabolites that were unidentified in this study. DAHP, 3-deoxy-D-arabino-heptulosonate-7-phosphate; PK, pyruvate kinase; PPH, phosphopyruvate hydratase; SAT, serine O-acetyltransferase; PSAT, Phosphoserine aminotransferase; CM, chorismate mutase; CS, chorismate synthase; DHD, 3-dehydroquinate dehydratase; DHQS, 3-dehydroquinate synthase; EPSPS, 3-phosphoshikimate 1-carboxyvinyltransferase; SDH, shikimate dehydrogenase; DSD, 3-dehydroshikimate dehydratase; ALT, alanine transaminase; AS, anthranilate synthase; Cyss, cysteine synthase; DAHPS, 3-deoxy-7-phosphoheptulonate synthase; SHMT, serine hydroxymethyltransferase; DLD, dihydrolipoyl dehydrogenase; DLAT, dihydrolipoyllysine-residue acetyltransferase; AspS, asparagine synthase; MDH, malic dehydrogenase (malate dehydrogenase); ACLY, ATP citrate synthase; CITS, citrate synthase; AH, aconitate hydratase; OGD, 2-oxoglutarate dehydrogenase (succinyl-transferring); DLST, dihydrolipoyllysine-residue succinyltransferase; SD, Succinate dehydrogenase; AK, aspartate kinase; ASADH, aspartate-semialdehyde dehydrogenase; HSDH, homoserine dehydrogenase; HSK, homoserine kinase; BCAT, branched-chain-amino-acid transaminase; NiR, ferredoxin-nitrite reductase; NR, nitrate reductase; GS, glutamate synthase; GOGAT, NAD(+)-dependent glutamate synthase; GDH, glutamate dehydrogenase; HDH, histidinol dehydrogenase; NAGS, N-acetylglutamate synthase; GSADH, glutamate-5-semialdehyde dehydrogenase; P5CR, pyrroline-5-carboxylate reductase; TS, theanine synthetase; ASS, argininosuccinate synthase; ASL, argininosuccinate lyase; GAD, glutamate decarboxylase. DHQD, 3-dehydroquinate dehydratase.