Figure 4.
Development of a Genetically Encoded Pyruvate Sensor Based on Fluorescence Resonance Energy Transfer
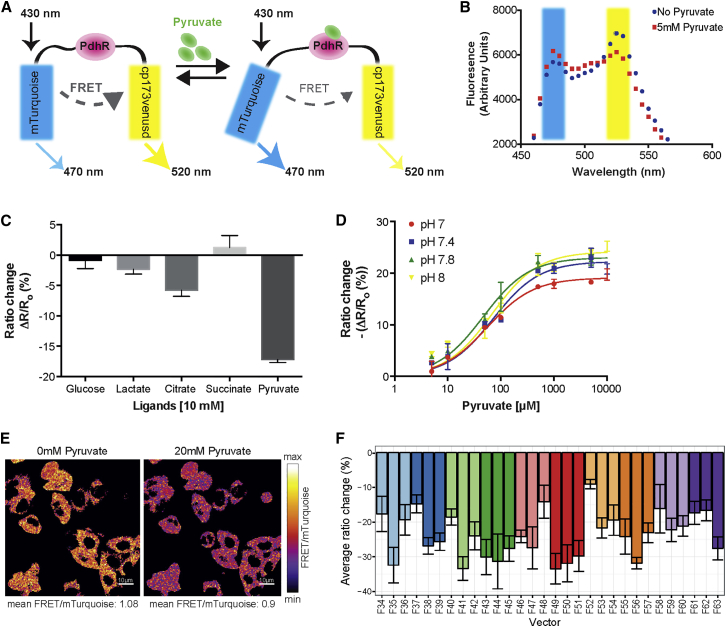
(A) Design of FRET-based sensor for pyruvate. E. coli pyruvate dehydrogenase regulator (PdhR) protein was used as the binding domain. Upon binding pyruvate, the domain undergoes a reversible conformational change, which results in a decrease in FRET from the mTurquoise donor to cp173Venusd acceptor.
(B) Fluorescence emission spectra of recombinant mTurquoise-PdhR-cp173Venusd protein after excitation at 430 nm in the presence (red dotted line) or absence (blue dotted line) of 5 mM pyruvate.
(C) Ligand specificity of the FRET sensor. Purified recombinant mTurquoise-PdhR-cp173-Venusd protein was excited at 430 nm in the presence or absence of different ligands (at 10 mM final concentration). Ratio (R) of FRET acceptor emission intensity to FRET donor emission intensity was calculated before (Ro) and after (Rf) ligand addition and the % ratio change ((Rf − Ro)/Ro) was obtained and plotted. Data are presented as mean ± SD. n = 3.
(D) Effect of pH on the FRET response of the pyruvate sensor. The effect of pH on the FRET response of recombinant mTurquoise-PdhR-cp173-Venusd protein was studied by incubating the protein for 30 min in the respective pH buffer (pH 7–8) and pyruvate was then titrated in the same buffer. Measurements were recorded and % ratio change (−(Rf − Ro)/Ro) was obtained. Data were fitted to one site binding equation using GraphPad Prism software and are presented as mean ± SD. n = 2.
(E) HeLa cells expressing mTurquoise-PdhR-cp173Venusd show drop in FRET ratio (Venus/mTurquoise fluorescence) upon addition of 20 mM pyruvate. Shown are the ratiometric images (Venus/mTurquoise fluorescence) of HeLa cells before and after addition of pyruvate.
(F) Optimization of pyruvate sensor by FRET library screening in HeLa cells. PdhR protein was cloned in between different variants of mTurquoise and Venus attached via 2, 4, or 8 amino acid linker length peptides. The mean ratio change for each construct in HeLa cells (24 cells average for each construct) upon addition of 20 mM pyruvate was quantified from averaged cell traces and plotted. Data are presented as mean ± SD.