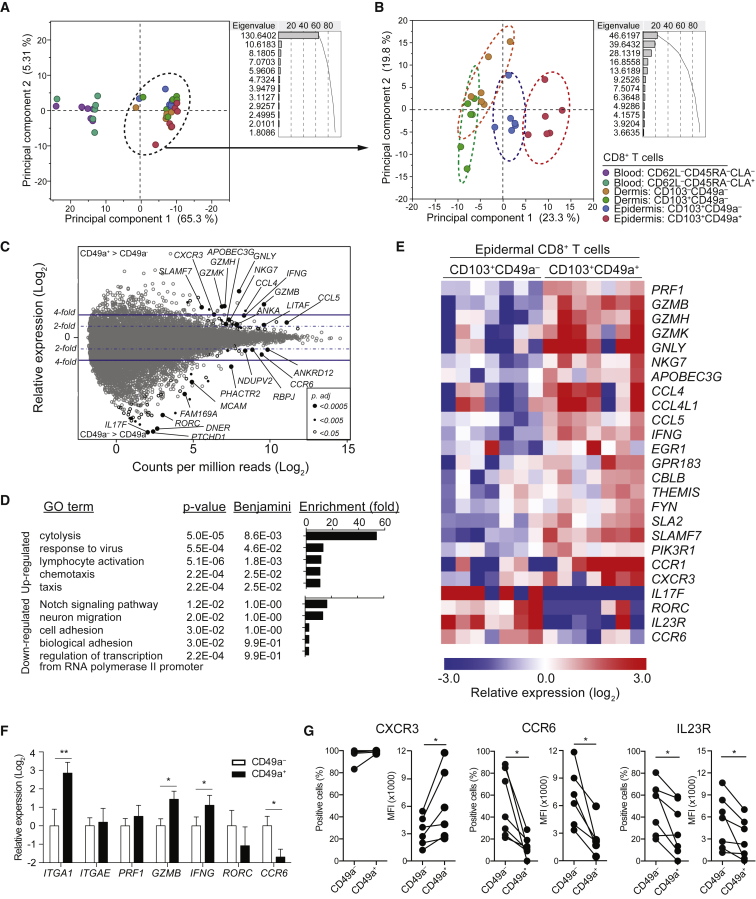
Figure 2.
Distinct Transcriptional Profiles of CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm Cells
(A–E) RNA-seq analyses of CD8+ T cell subpopulations sorted from blood, dermis, or epidermis of healthy individuals. (A and B) Principal component analysis of the top 200 differentially regulated genes among (A) all six subpopulations or (B) four skin-derived subpopulations after mean centering to correct for batch effects. Dots indicate samples of different subpopulations from a total of 6 donors, as indicated. Principal component 1 and 2 represents the largest source of variation, combined accounting for (A) 70.6% and (B) 43.1% of the total variation, respectively. Color-coded eclipses indicate the 95% confident area of particular T cell subpopulations. (C–E) Transcriptome analysis of CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cells sorted from healthy epidermis (n = 7). (C) MA-plot (Log2 fold change against Log2 count per million) showing differential gene expression between epidermal CD8+CD103+CD49a− and CD8+CD103+CD49a+ Trm cells. Each gene was symbol-coded as indicated according to their adjusted p values generated using EdgeR with FDR correction. (D) Functional annotation analysis using DAVID tool listing gene sets upregulated (upper) or downregulated (lower) of CD8+CD103+CD49a+ Trm cells as compared to CD8+CD103+CD49a– Trm cells. (E) Heatmap showing gene expression of selected genes associated with functional annotations. Each column represents an individual donor. Row-mean centered relative expression values are shown.
(F) Quantitative real-time PCR measurements of transcript levels of specific genes, as indicated, in epidermal CD8+CD103+CD49a– relative to CD8+CD103+CD49a+ Trm cell subsets. Fold-change was calculated against the mean expression of the CD8+CD103+CD49a– Trm cells. Mean ± SD depicted.
(G) Surface expression of CXCR3, CCR6 and IL-23 receptor (IL23R) in CD8+CD103+CD49a– or CD8+CD103+CD49a+ Trm cells, as assessed by flow cytometry. Data are presented with respect to frequency of positive cells and their respective median florescence intensity (MFI) in subsets, as indicated. Two-tailed Wilcoxon tests. ∗p < 0.05, ∗∗p < 0.01. See also Figure S1.