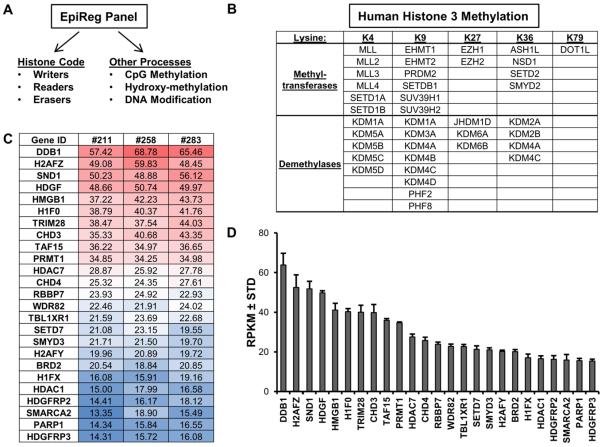
Figure 2. Robustly expressed epigenetic regulators in MSCs.
The major classes of human epigenetic regulators (A). The histone code can be modified and interpreted by several classes of genes, including enzymes that add (writers: e.g., methyltransferases, acetyltransferases) or remove (erasers: e.g., demethylases, deacetylases) functional groups on histones. Other proteins have domains (readers: e.g., bromodomain, chromodomain) that bind to specific histone modifications and control gene expression by recruiting other transcriptional modulators. Methylation of H3 on K4, K9, K27 or K36, which represents a critical epigenetic post-translational modification that controls gene expression, is modulated by several methyltransferases and demethylases (B). The table (C) and graph (D) depict the expression patterns of the 25 most robustly expressed epigenetic regulators in non-proliferative (confluent) MSCs (n = 3). The table and graph show expression values expressed as reads per kilobasepair per million mapped reads (RPKM).