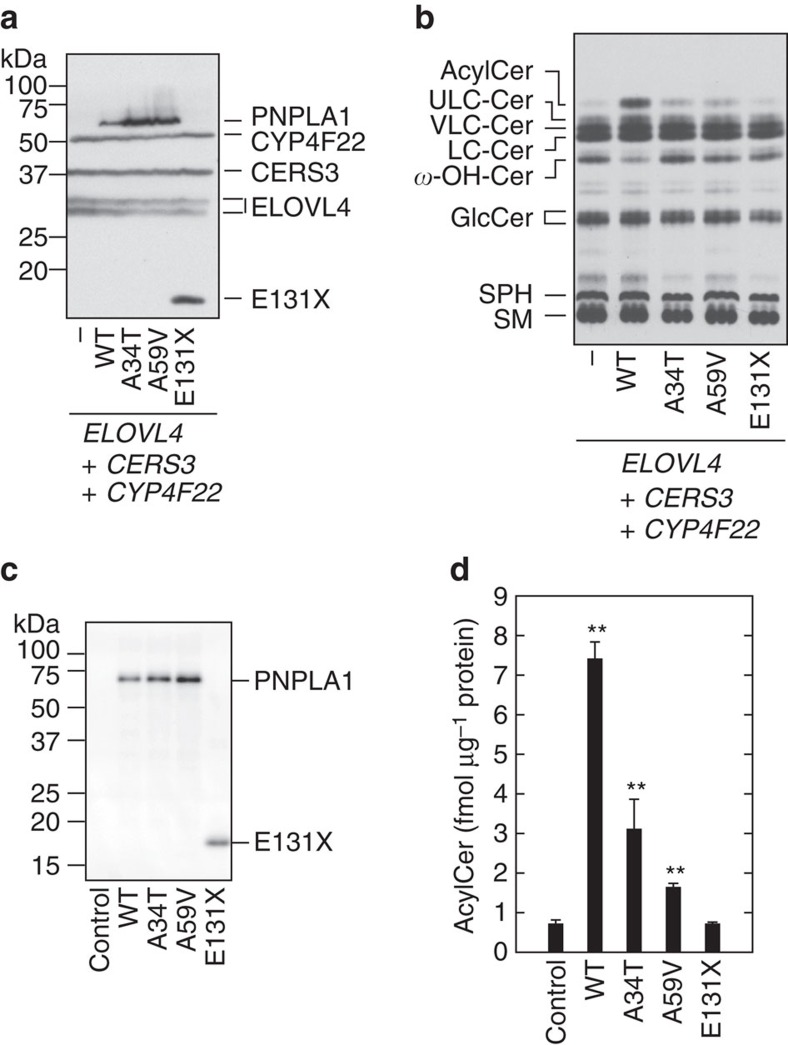
Figure 6. PNPLA1 activity is impaired in ichthyosis-causative mutants.
(a,b) HEK 293T cells were transfected with the plasmids encoding 3xFLAG-ELOVL4, 3xFLAG-CERS3, 3xFLAG-CYP4F22 and 3xFLAG-PNPLA1 (wild type (WT) or mutant) as indicated. (a) Total cell lysates were separated by SDS-PAGE and subjected to immunoblotting with anti-FLAG antibody. (b) Cells were labelled with [3H]sphingosine in the presence of 25 μM linoleic acid for 6 h at 37 °C. Lipids were extracted, separated by normal-phase TLC and detected by autoradiography. AcylCer, acylceramide; Cer, ceramide; GlcCer, glucosylceramide; SM, sphingomyelin; SPH, sphingosine; ω-OH-Cer, ULC ω-hydroxyceramide. Three independent experiments gave similar results, and a representative result is shown. (c,d) Genes on control plasmid (pEU-E01-T1R1) or plasmid encoding wild type (WT; pEU-E01-3xFLAG-PNPLA1) or mutant PNPLA1 (for A34T, pEU-E01-3xFLAG-PNPLA1(A34T); for A59V, pEU-E01-3xFLAG-PNPLA1(A59V); or for E131X, pEU-E01-3xFLAG-PNPLA1(E131X)) were transcribed in vitro using SP6 RNA polymerase. The resulting mRNAs were then incubated with wheat germ lysates and phosphatidylcholine-based liposomes containing TG and C30:0 ω-hydroxyceramide. (c) Proteins associated with the resulting proteoliposomes were separated by SDS-PAGE, followed by immunoblotting with anti-FLAG-antibody. (d) An in vitro acylceramide synthesis assay was performed by incubating proteoliposomes for 1 h at 37 °C. Lipids were extracted, and the C30:0 acylceramide EOS was quantified by LC-MS analysis. Values represent the means±s.d.s of three independent experiments. Statistically significant differences compared to control are indicated (two-tailed Student's t test; **P<0.01).