Figure 2.
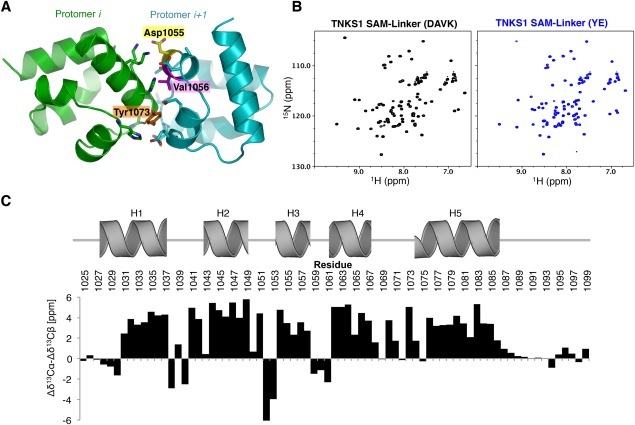
SAM mutants retain a folded structure. (A) Residues at the interface of two protomers in the predicted oligomer interface of TNKS1. Residues that were mutated to disrupt oligomerization are highlighted. (B) 1H15N‐HSQC NMR analysis of the TNKS1 SAM‐Linker(DAVK) mutant (left), and the TNKS1 SAM‐Linker(YE) mutant (right). DAVK, D1055A/V1056K; YE, Y1073E. Both spectra are well dispersed and highly similar, consistent with folded, monomeric domains. (C) Chemical shift differences between random coil and experimentally determined Cα and Cβ atoms (Δδ13Cα‐Δδ13Cβ [ppm]). Positions of Rosetta‐predicted helices are shown above histogram. High values are predictive of helical structure.
