Abstract
Apposite development of anther and its dehiscence are important for the reproductive success of the flowering plants. Recently, bHLH142, a bHLH transcription factor encoding gene of rice has been found to show anther-specific expression and mutant analyses suggest its functions in regulating tapetum differentiation and degeneration during anther development. However, our study on protein level expression and gain-of-function phenotype revealed novel aspects of its regulation and function during anther development. Temporally dissimilar pattern of bHLH142 transcript and polypeptide accumulation suggested regulation of its expression beyond transcriptional level. Overexpression of bHLH142 in transgenic rice resulted in indehiscent anthers and aborted pollen grains. Defects in septum and stomium rupture caused anther indehiscence while pollen abortion phenotype attributed to abnormal degeneration of the tapetum. Furthermore, RNA-Seq-based transcriptome analysis of tetrad and mature pollen stage anthers of wild type and bHLH142OEplants suggested that it might regulate carbohydrate and lipid metabolism, cell wall modification, reactive oxygen species (ROS) homeostasis and cell death-related genes during rice anther development. Thus, bHLH142 is an anther-specific gene whose expression is regulated at transcriptional and post-transcriptional/translational levels. It plays a role in pollen maturation and anther dehiscence by regulating expression of various metabolic pathways-related genes.
Major events in the anther development are differentiation of stamen primordia, development of microspore and then dehiscence of the anther. Typical anther has four locules and each locule consists of four-layered anther wall in which the developing microspore resides1,2. Each layer of the anther wall performs specialised function during the process of anther development3. Epidermis is the outer cover that protects anther from various environmental stresses and also forms specialised structures named as stomium and septum, which are involved in anther dehiscence process4,5. Endothecium is the second layer that develops secondary thickening in the form of lignin deposition, which assists the process of anther dehiscence. Middle layer is present between endothecium and tapetum, which undergoes degeneration during pollen maturation along with the tapetum. Tapetum is the innermost layer, which undergoes programmed cell death (PCD)-mediated degeneration to release nutritive components for developing pollen and sporopollenin as well as other pollen wall precursors6. Proper development and timely degeneration of specific cell types in the anther wall layer is essential for development and dispersal of pollen grains7,8,9. Various cellular and metabolic changes occur during the formation and degeneration of each layer. In recent years, role of bHLH transcription factors in various aspects of anther development has been elucidated. DYSFUNCTIONAL TAPETUM1 (DYT1) from Arabidopsis and its rice homolog UNDEVELOPED TAPETUM1 (UDT1) reportedly regulate the tapetum development process. Mutant plants of both the genes displayed hypertrophic growth and abnormal vacuolation of tapetum10,11,12. Various lipid metabolism, cell-wall modification and secondary metabolism-related genes were downregulated in the dyt1 mutant12. Furthermore, bHLH10, bHLH89 and bHLH91 have been shown to interact with DYT1 and work redundantly during anther development13. Similarly, mutation in the ABORTED MICROSPORE1 (AMS1) gene in Arabidopsis, and its rice ortholog, TAPETUM DEGENERATION RETARDATION (TDR) result in pollen abortion and a hypertrophic tapetum14,15. AMS interacts with bHLH89 and bHLH91 and acts as a master regulator of pollen wall development by directly regulating expression of genes related to various metabolic processes16,17. TDR affects the metabolism of fatty acids and other aliphatic compounds besides regulating tapetum degeneration by directly regulating the expression of a cysteine protease gene CP114,18. Furthermore, another bHLH transcription factor, ETERNAL TAPETUM1 (EAT1) has been shown to interact with TDR and regulates tapetal PCD in rice19. Recently, mutant analyses of bHLH142 from rice revealed its role in tapetum differentiation and degeneration during post-meiotic anther development20,21.
Modified epidermal tissues including septum and stomium along with endothecium are involved in the process of anther dehiscence22. A number of genes that are implicated in the process of anther dehiscence have been identified from forward and reverse genetics studies in Arabidopsis and rice23. DEFECTIVE IN ANTHER DEHISCENCE1 (DAD1) gene encodes phospholipase enzyme required for jasmonic acid biosynthesis and regulates anther dehiscence in Arabidopsis. Knock-down/Knock-out of SIZ1, a SUMO E3 ligase gene, resulted in sterile rice plant due to lack of anther dehiscence24. Pollen Semi-Sterility1 (PSS1) encodes a kinesin-like protein that regulates both pollen development and anther dehiscence process in rice25. A rice MYB transcription factor encoding gene ANTHER INDEHISCENCE1 (AID1) has been shown to be involved in anther dehiscence process by regulating septum and stomium degradation26. A mutation in another MYB gene MALE STERILE35 (MYB26) from Arabidopsis leads to male sterility because of anther indehiscence27,28. Furthermore, MYB26 has been found to regulate secondary thickening of endothecium by affecting the expression of genes related to lignin deposition in secondary walls27. Thickening of the endothecium secondary wall in Arabidopsis anther is known to be regulated by NAC transcription factors NST1 and NST229.
Previously, we had characterised the promoter of bHLH142 through transgenic approaches and shown its capability to impart anther-specific expression to the reporter gene30. In this study, we show that although the bHLH142 transcript accumulation peaks during early stages of anther development the resultant protein accumulates in a biphasic manner; once at the tetrad stage of the anther and then in mature anther, in spite of relatively low levels of its transcript being present in the mature anther. Phenotypic as well as transcriptome analysis of bHLH142OE transgenic plants revealed that bHLH142 regulates anther dehiscence and tapetum degeneration process by affecting cell wall degradation and ROS signalling-related genes and it controls pollen maturation by affecting carbohydrate and lipid metabolism-related genes.
Results
bHLH142 shows biphasic expression pattern at protein level
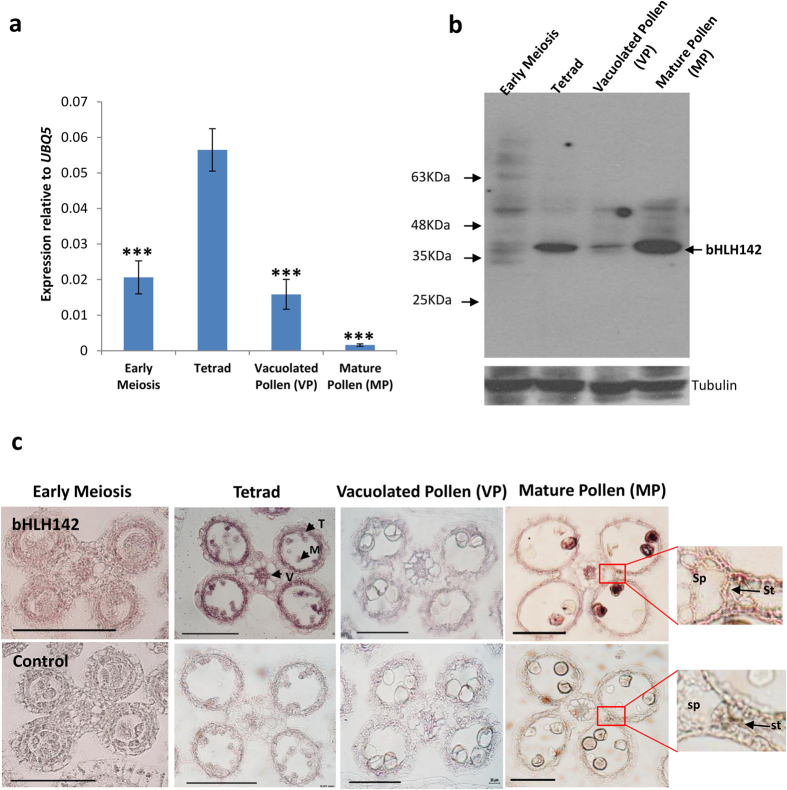
In a previous report, we showed that bHLH142 is an anther-specific gene in rice and its promoter displayed maximum activity in meiotic anther30. Later, RNA-in situ hybridisation studies by Fu et al.21 and Ko et al.20 also confirmed that bHLH142 transcripts accumulate predominantly at the meiotic anther stage. The accumulation of bHLH142 mRNA and its protein during different stages of anther development was analysed to gain more insight into its expression pattern by qRT-PCR (Quantitative Real Time-PCR) and immunoblotting techniques, respectively. Consistent with the previous reports, bHLH142 mRNA was detected in early meiosis stage and showed maximum abundance in tetrad stage. After these stages, the transcript levels declined in the vacuolated pollen (VP) stage and were barely detectable in the mature pollen (MP) stage of anther development (Fig. 1a). Furthermore, bHLH142 protein sequence analysis showed two bipartite and three monopartite nuclear localization signals, which suggested its possible nuclear localization (Figure S1). To confirm this, subcellular localization study was performed and result showed that bHLH142-YFP fusion protein localised exclusively into the nucleus (Figure S1). Furthermore, accumulation of bHLH142 protein during different stages of rice anther development was examined through immunoblotting. A faint band of bHLH142 protein was detected at early meiosis stage and vacuolated pollen stage but a rather strong expression was observed at tetrad and MP stage (Fig. 1b). This suggests that bHLH142 protein accumulated in tetrad as well as in MP stage of the anther. Detection of bHLH142 protein in MP stage was surprising as neither a significant promoter activity nor the transcript was reported at this stage20,21,30. Consistent results from three independent experiments confirmed the presence of bHLH142 polypeptide in the MP stage of rice anther. To rule out that the hybridisation with other bHLH proteins could give false signals in mature anthers, the anti-bHLH142 antibody was tested for cross reactivity with two of its closest homologues, EAT1 and TDR. The absence of any cross-reactivity proved accumulation of bHLH142 in mature anthers (Figure S1). To investigate distribution of bHLH142 protein in different cell types of anther during different stages and to substantiate its detection at MP stage, in situ immunolocalization study was performed. Outcome supported the immunoblot data, as a faint signal was detected in anther cross-sections from early meiosis stage and vacuolated pollen stage compared to strong signal in tetrad and MP stage (Fig. 1c). Furthermore, bHLH142 polypeptide was detected in the tapetum and microspores during tetrad stage and in epidermis and pollen grains at MP stage. Enlarged view of the wall layers of MP anther showed presence of immune signal in the septum and stomium regions. In addition, expression signal was also detected in vascular tissue of tetrad and MP stage of the anther. Control samples showed only background signals (Fig. 1c). Therefore, cell type-specific distribution of bHLH142 mRNA20,21 and the polypeptide appeared similar, as both were detected in tapetum, microspores and vascular tissues. However, the levels of the transcript and the polypeptide varied during different anther stages, as although the mRNA was present in negligible amounts at the MP stage, the protein accumulated in high amounts. This suggests that there is temporal difference in expression pattern of bHLH142 transcript and polypeptide while spatial pattern is similar.
Figure 1. Temporal expression pattern of transcript and protein for bHLH142 are different.
(a) qRT-PCR expression analysis of bHLH142 transcript in different stages of rice anthers. Error bars indicate standard deviation (SD). The data are presented as the mean ± SD (n = 3). Asterisks indicate significant difference with respect to Tetrad (‘***’ indicates t-test, p-value ≤ 0.001). (b) Immunoblot showing accumulation of bHLH142 polypeptide in different stages of rice spikelets. (c) In situ immunolocalization showing spatio-temporal presence of bHLH142 polypeptide during different stages of rice anthers. Right most panels show enlarged view of the selected region (shown as rectangle) of MP stage of the anther. Control panel shows samples without bHLH142 primary antibody treatment. M, Microspore; sp, Septum; st, Stomium; T, Tapetum; V, Vascular tissue. Scales: 100 μm.
Overexpression of bHLH142 causes defects in degeneration of septum and stomium that leads to indehisced anther
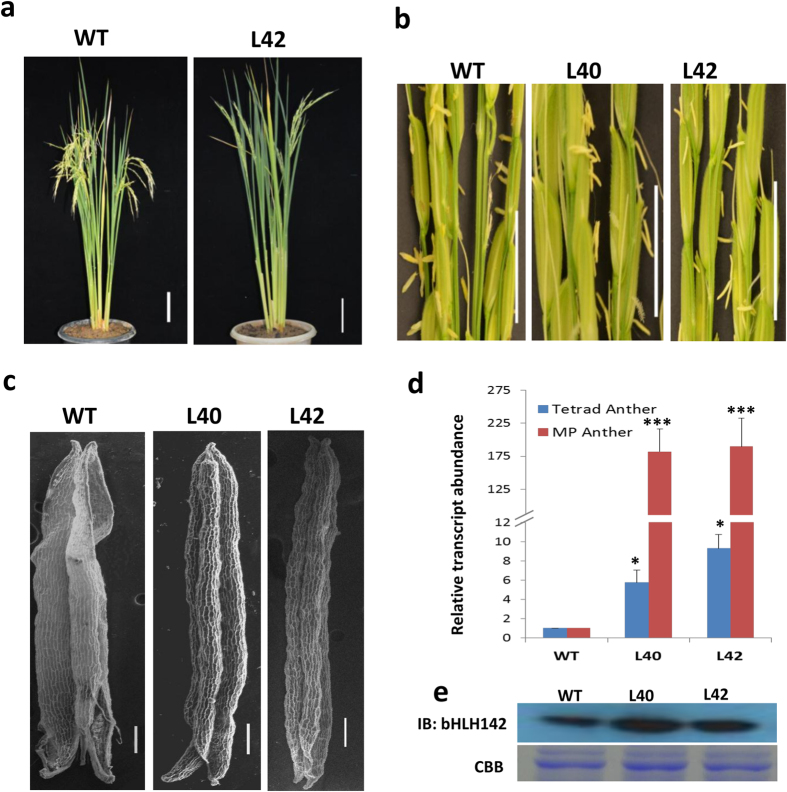
Detection of bHLH142 protein in biphasic manner at tetrad and MP stage anthers raised the possibility of its function during both the stages. However, recent studies of bHLH142 mutants suggested its role only during tetrad stage in controlling tapetum development/degeneration20,21. Functional characterisation of a gene by gain-of-function approach helps in exploring its novel functions during plant growth and development31,32,33,34. Therefore, we used gain-of-function approach to get more insight into biological functions of bHLH142 during rice anther development. Rice transgenics overexpressing bHLH142 under the control of a maize ubiquitin promoter were raised (Figure S2 and Table S1). At least seven independent positive transgenic plants showed high level of bHLH142 transcript accumulation (Figure S2). All of them showed normal vegetative growth, but none of them was able to set seeds. However, manual pollination of transgenic plants with wild type (WT) pollen resulted in seed formation (Figure S3). This suggested that defects in the male reproductive development affected the seed set. Seeds obtained from cross pollination were grown and resultant plants showed segregation of hygromycin-resistant marker gene into 1:1 ratio (Figure S3). Positive T1 plants were again sterile and not able to form seeds despite having normal vegetative growth (Fig. 2a). Observation of the male reproductive organs in WT and transgenic plants revealed that anthers of the transgenic plants did not dehisce even after the complete maturation (Fig. 2b). All analyses were performed in these T1 transgenic plants. However, to look at the stability of phenotype, we observed one more generation and each time similar results were obtained, suggesting that the phenotype was stably inherited in transgenic plants (Figure S3).
Figure 2. Overexpression of bHLH142 causes anther indehiscence in rice.
(a) bHLH142OEplant (L42) showing growth compared to WT. (b) Post-anthesis panicles of WT and bHLH142OE transgenics (L40 and L42) showing dehisced anthers in WT and indehisced anther in transgenic plants. (c) SEM images of WT and transgenic anthers showing that apical and basal part of WT anther is ruptured while that of transgenics is intact. (d) qRT-PCR expression analysis showing high accumulation of bHLH142 transcripts in transgenic anthers at tetrad and MP stages. Error bars indicate standard deviation (SD). The data are presented as the mean ± SD (n = 3). Asterisks indicate significant difference with respect to WT (‘*’ indicates t-test p-value ≤ 0.05 while ‘***’ indicates p-value ≤ 0.001). (e) Immunoblot showing higher accumulation of bHLH142 polypeptide in transgenic plants in MP anther. WT, Wild Type.
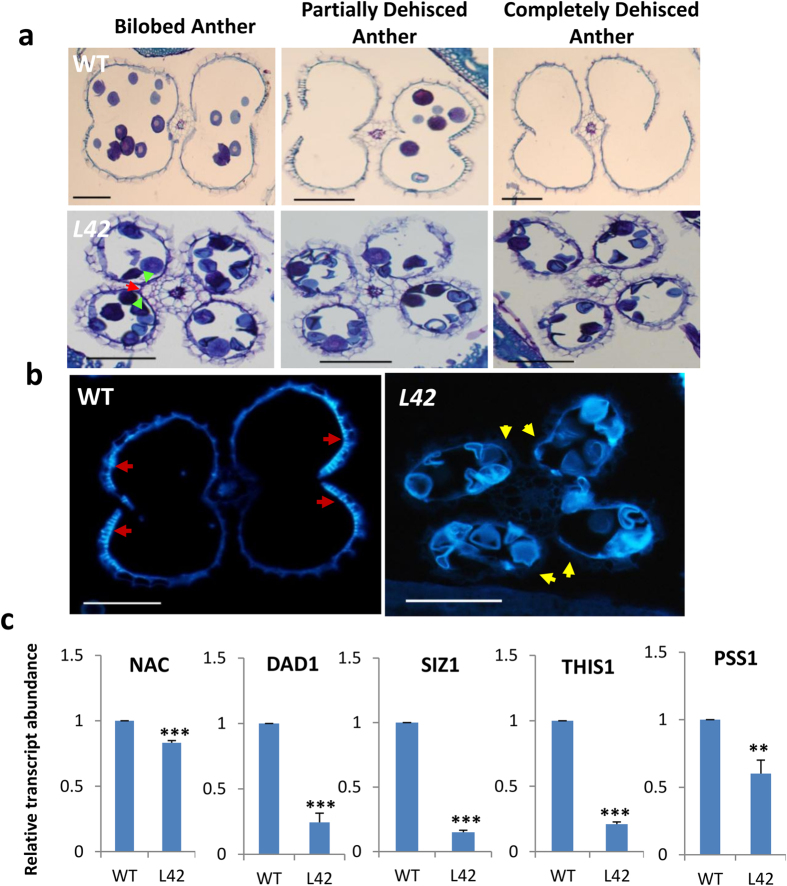
We compared the anther dehiscence phenomenon in the transgenics with that in the WT. Scanning electron microscopic observations of the post-anthesis-staged anthers revealed that the apical and basal parts of the transgenic anthers remained intact while they were found to be ruptured in WT (Fig. 2c). bHLH142 transcripts as well as polypeptide were highly overexpressed in bHLH142OEtransgenic plants compared to WT as analysed by qRT-PCR and immunoblotting (Fig. 2d,e). Furthermore, observation of transverse sections of WT and transgenic spikelets at mature stage showed that anther dehiscence in WT was initiated by degradation of septum tissue, which causes two anther locules to fuse together. Then another set of specialised cells forming stomium at the junction of two locules ruptured to complete the process of dehiscence and allow the release of pollen grains (Fig. 3a). However, bHLH142OE anther locules did not fuse together owing to intact septum and also stomium did not rupture, preventing the release of pollen grains from the anther (Fig. 3a). Moreover, in WT plants, thickening of endothecium as band-like structure near the junction of the two locules was observed in mature anther undergoing dehiscence but such structure was absent in transgenic anthers (Fig. 3a). To confirm this, mature anther sections were analysed under UV light in fluorescence microscope and endothecium thickenings due to lignin deposition were found to be absent in transgenic anthers (Fig. 3b). Furthermore, we checked the expression of genes associated with anther dehiscence in rice and found that DAD1, SIZ1 and THIS1 were downregulated in the bHLH142OE anthers (Fig. 3c).
Figure 3. bHLH142 overexpressing anthers are defective in septum and stomium degeneration.
(a) Transverse sections of pre-anthesis staged WT and transgenic anthers (L42) undergoing dehiscence. Green and red arrows indicate intact septum and stomium in transgenic anthers, respectively. (b) WT and transgenic anthers analyzed under UV light to visualize lignin autofluorescence of endothecium regions. Red and yellow arrows indicate presence and absence secondary thickening of endothecium, respectively. (c) qRT-PCR expression data of anther dehiscence-related protein genes. Error bars indicate standard deviation (SD). The data are presented as the mean ± SD (n = 3). Asterisks indicate significant difference with respect to WT (‘***’ indicates t-test p-value ≤ 0.001, while ‘**’ indicates p-value ≤ 0.01). WT, Wild Type. Scales: A, 50 μm; B, 100 μm.
bHLH142 overexpressing anther produces defective and aborted pollen grains
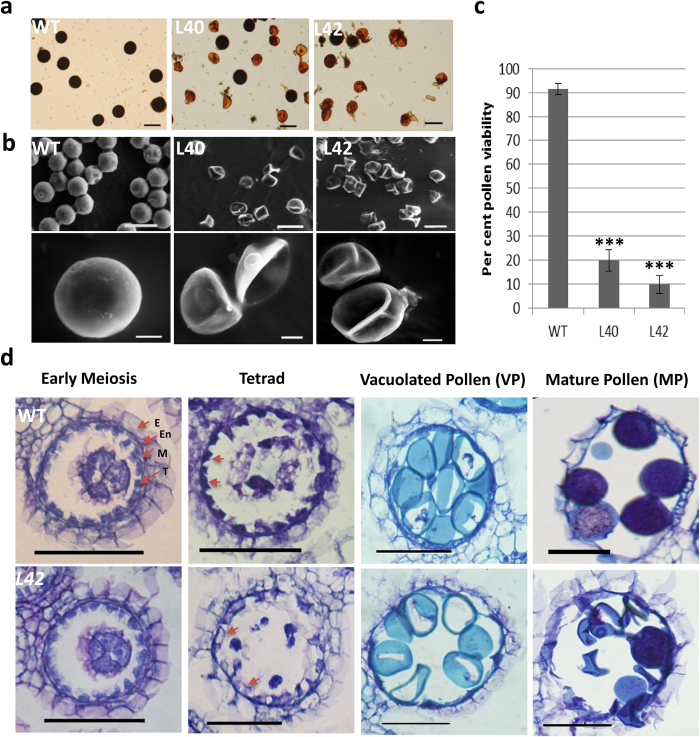
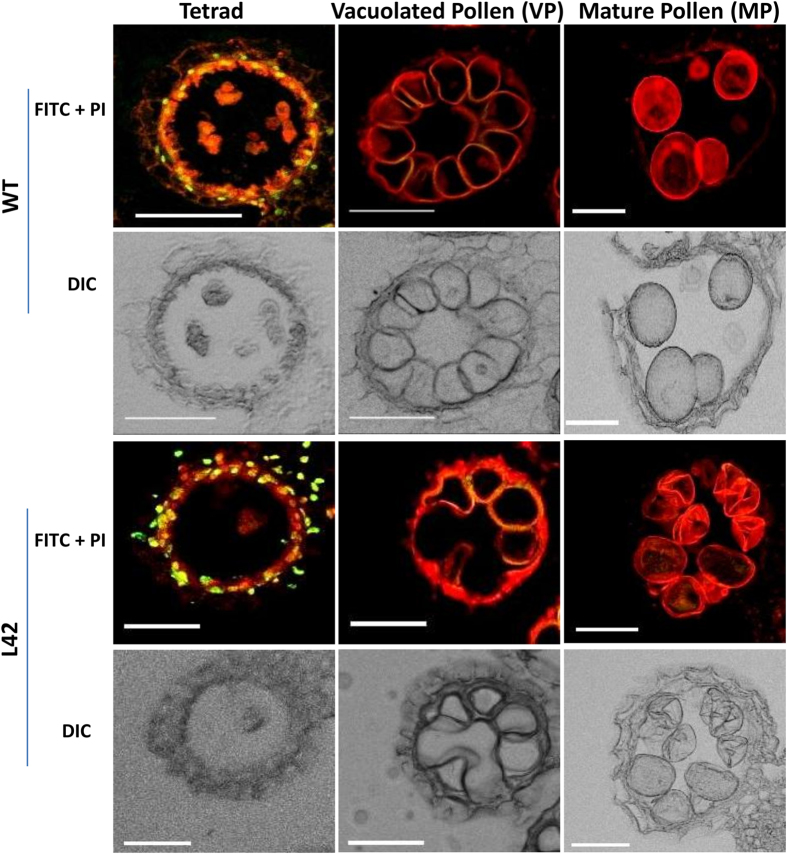
To explore the male reproductive development in bHLH142OE plant we analysed its pollen development process. Most of the pollen grains of transgenic plants were found to be non-viable as they did not take up I2-KI stain (Fig. 4a). Electron microscopic analysis showed that transgenic pollen grains were shrunken and abnormal in shape when compared to WT (Fig. 4b). We tested pollen viability in several transgenic lines in T0, T1 and T2 generations and all showed high reduction in the viability (Figs 4c and S3). However, viable pollen grains showed normal pollen germination similar to WT (Figure S3) and manually dehisced anthers were able to cause set seed. This suggests that overexpression of bHLH142 affects pollen development but surviving pollen grains have ability to fertilize the female gametes. To explore the pollen development process in transgenics, paraffin sections of the WT and bHLH142OE anthers at different stages of development were analysed. No developmental differences were observed during early meiosis stage. All the four anther layers namely; epidermis, endothecium, middle layer and tapetum appeared to be differentiated and enlarged, nucleated microspore mother cells were also seen. At tetrad stage, WT anther had well organised deeply stained tapetal cell layer while transgenic anther tapetal cells appeared distorted and poorly stained (Fig. 4d). It also appeared that tapetum had loosely filled cytoplasm compared to that in the WT. However, no difference in microspore development was observed till vacuolated pollen stage (Fig. 4d). However, at MP stage most of the transgenic microspores failed to mature and eventually aborted, while WT pollen were filled with starch and attained maturity. These observations suggested some alteration in tapetum development in transgenic plants that probably restricted the pollen maturation process. It is known that tapetum undergoes PCD-mediated degeneration after the meiosis stage to nourish the developing microspore/pollen grain6,35. To identify the reason behind appearance of the abnormal tapetal cells in transgenic anther, tapetum degeneration process in WT and transgenic anthers was studied through TUNEL (Terminal deoxynucleotidyl transferase dUTP Nick End Labeling) assay. In the WT anthers, positive TUNEL signal was observed only in tapetal cells of tetrad stage anther and not from vacuolated and mature pollen anther (Fig. 5). In bHLH142OE anther also, TUNEL signal was detected only at tetrad stage but, intensity of the signal as well as the number of tapetal cells showing the signal was much more as compared to the WT. This suggested that bHLH142 overexpressing transgenic anthers probably undergo faster tapetal degeneration compared to wild type anthers (Fig. 5). Hence, abnormal tapetum in transgenic anther may have appeared because of aberrant degeneration, which affected maturation of vacuolated microspores into mature pollen grains due to the lack of proper nutrition.
Figure 4. bHLH142 overexpressing anthers produce defective pollen grains.
(a) I2-KI staining of pollen grains from WT and transgenic plants (L40 and L42). (b) SEM images of WT and transgenic pollen grains (L40 and L42). (c) Quantitative estimation of pollen viability through I2-KI staining. Error bars indicate standard deviation (SD). The data are presented as the mean ± SD (n = 3). Asterisks indicate significant difference with respect to WT (‘***’ indicates t-test, p-value ≤ 0.001). (d) Transverse sections of WT and transgenic anthers (L42) during various stages of development. Arrows in the tetrad panel indicate well developed and poorly developed tapetum in WT and transgenic anthers, respectively. WT, Wild Type; E, Epidermis; En, Endothecium; M, Middle layer; T, Tapetum. Scales: (a), 50 μm; (b), upper panel 50 μm, lower panel 10 μm; (d), 50 μm.
Figure 5. bHLH142 overexpressing anther shows abnormal programmed cell death in tapetum.
TUNEL assay showing PCD undergoing cells as greenish-yellow signal due to merging of FITC signals and propidium iodide (PI) stain and normal cells appear red due to propidium iodide stain only. Images were taken from confocal microscope in FITC (Fluorescein isothiocyanate), PI and DIC (Differential interference contrast) channels. Merge image of FITC and PI are shown in fluorescent panel while DIC images are shown separately. Scales: 50 μm.
bHLH142 regulates cell wall degradation, carbohydrate and lipid metabolism and ROS homeostasis-related genes during rice anther development
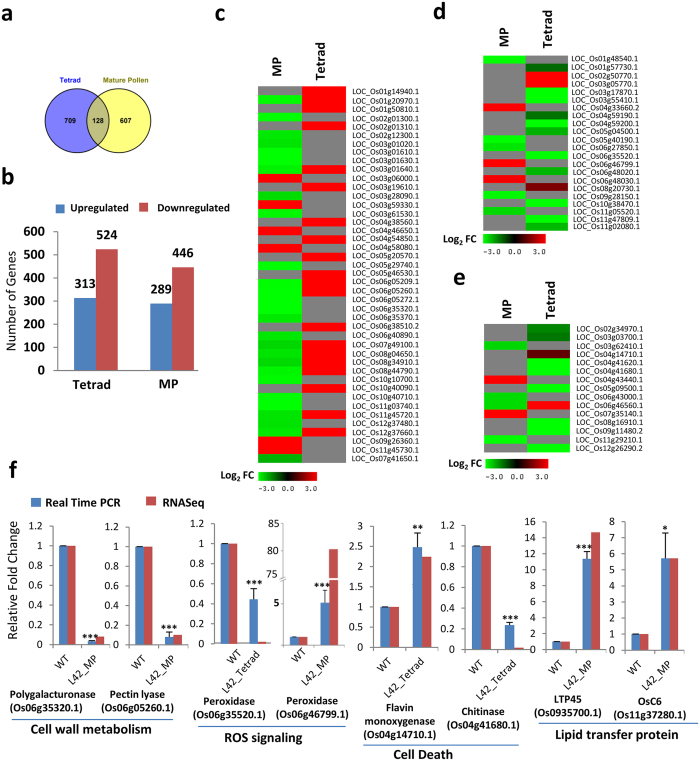
Transcription factors perform their biological functions through regulating expression of various signalling and metabolic pathways-related genes. Maximum expression of bHLH142 protein was detected at tetrad and MP stage of the rice anthers. Therefore, to identify the downstream gene regulatory network of bHLH142, RNA-Seq-based transcriptome analysis of tetrad and MP stage of WT and bHLH142OE anthers was performed. Differentially expressed genes in both the stages of transgenic and WT anthers were identified. Total 837 genes in tetrad and 735 in MP anther were found to be differentially expressed in bHLH142OE compared to WT. However, only 128 genes were found to be common in both the stages (Fig. 6a). In tetrad anther, 313 genes were upregulated and 524 were downregulated. In MP anther, 289 upregulated and 446 downregulated genes were identified (Fig. 6b). In order to classify these genes into functional categories, Gene Ontology (GO) terms were identified and their enrichment was performed. Furthermore, putative functions of each gene were retrieved from RGAP (Rice Genome Annotation Project). Regulation of anatomical structure, cell growth, cell size and cellular component size-related GO terms were enriched in differentially expressed genes at tetrad stage. Carbohydrate metabolic process, reproduction, enzyme and hydrolytic activity terms were enriched in MP stage (Figure S4).
Figure 6. bHLH142OE anthers show changed in expression of various metabolic pathway-related genes.
(a) Venn diagram showing common and unique genes differentially regulated in tetrad and MP anther. (b) Histogram showing number of genes upregulated and downregulated in tetrad and MP stage of transgenic anthers compared to WT. Heat map showing log2 fold changed in expression of cell wall modification (c), ROS signaling (d) and cell death (e)-related genes. (f) Validation of changed in expression of different metabolism-related genes through qRT-PCR. The data are presented as the mean ± SD (n = 3). Asterisks indicate significant difference with respect to WT (‘***’ indicates t-test p-value ≤ 0.001, ‘**’ p-value ≤ 0.01 and ‘*’ p-value ≤ 0.05).
Furthermore, few of the key genes already known for anther development were also found to be differentially expressed in bHLH142OE plants (Table 1). Among these, OsC4, OsC6, CYP704B2 and CYP703A3 are known to be involved in pollen wall formation36,37,38,39 and MST8, INV4, SUT3, UGP2 and AGPL1 are known for sugar partitioning in reproductive tissues13,40. Moreover, genes belonging to different molecular and biological categories were found to be differentially expressed in bHLH142OE plants in tetrad and MP anthers (Tables S2 and S3). By considering GO and putative function obtained through RGAP, it was observed that genes related to carbohydrate metabolism, lipid metabolism, cell wall modification, ROS homeostasis and cell death represent major categories among differentially expressed genes in bHLH142 overexpressing anthers. Thirty-two genes in tetrad and thirty-eight genes in MP stage were found to be related with carbohydrate metabolism. Glycerophosphoryl diester phosphodiesterase, glycosyl transferase, polygalacturonase, glucan endo-1,3-beta-glucosidase and glycosyl hydrolase encoding genes were found to represent major carbohydrate metabolism-related differentially expressed genes (Table S4). Similarly, 23 genes in tetrad and 27 in MP were related to lipid metabolism. Among the lipid metabolism-related genes glycerophosphoryl diester phosphodiesterase, GDSL-like lipase/acylhydrolase genes were overrepresented (Table S5). Heat map showing expression of carbohydrate and lipid metabolism suggests that most of the genes are downregulated in both stages of anther (Figure S5). Furthermore, we obtained eight downregulated and two upregulated lipid transfer protein (LTP) encoding genes in tetrad stage and ten upregulated LTP genes in MP anther (Table S5). OsC4, OsC6, LTPL45 and CYP703A3 are notable LTP and lipid metabolism-related genes upregulated in MP stage, whose function in pollen development are already reported36,37,38. Changed expression of carbohydrate and lipid metabolism-related genes suggests that deposition of carbohydrate and lipid during pollen development is affected, which leads to formation of shrunken pollen in bHLH142OE plants. Besides carbohydrate and lipid metabolism, genes involved in cell wall modification, ROS signalling and cell deaths were found to be other overrepresented classes in transcriptome data. Thirty genes in MP stage were found to be related to cell wall modification and majority of them were downregulated. Also, 23 genes from this category were found in tetrad stage and most of them were upregulated (Fig. 6c). Genes encoding for enzymes related to pectin degradation and modification like, pectate lyase, pectinesterase, pectin methylesterase, polygalacturonase and expansin were found to be overrepresented among cell wall modification-related genes. (Table S6). Defect in stomium and septum lysis and formation of aborted pollen in bHLH142OE anther may be associated with down regulation of cell wall-related genes. Reactive oxygen species (ROS) signalling has been reported to play an important role during various growth and development processes. In the bHLH142OE anthers, fourteen ROS homeostasis-related genes in tetrad and eight in MP anther were found to be differentially expressed (Fig. 6d). ROS signalling-related genes showing differential expression mainly include peroxidases. However, metallothionin, thioredoxin monodehyrdroscorbate and gultathione-s-transferase were also found to be differentially expressed (Table S7). Furthermore, we obtained ten genes in tetrad and six genes in MP stage that were assigned GO-term ‘cell death’ (Fig. 6e and Table S7). Changed expression of cell death-related genes in tetrad stage may be associated with the defective tapetum degeneration, while in MP, it may have altered septum and stomuim degenerations. Changes in expression of several genes from different categories were validated through qRT-PCR (Fig. 6f). Histological studies of bHLH142OE plants showed that deposition of lignin was affected in transgenic anthers. We observed downregulation of some lignin biosynthesis-related genes in tetrad anther, which includes cinnamoyl CoA reductase, laccase and phenylalanine ammonia lyase (PAL) encoding genes. Furthermore, few water transporter protein aquaporin encoding genes were also downregulated in MP anthers (Table S8). Therefore, transcriptome analysis of bHLH142OE tetrad and MP anthers revealed that overexpression of bHLH142 transcription factor may lead to disturbance of metabolic processes like lipid and carbohydrate metabolism, cell wall modification, ROS homeostasis and cell death-related processes.
Table 1. List of previously known anther development and dehiscence-related genes showing differential expression in bHLH142 OE anthers compared to WT.
| Locus ID | Name | Putative Function | Log2 FC | p-Value | Regulation |
|---|---|---|---|---|---|
| LOC_Os01g38670.1 | MST8 | transporter family protein, putative, expressed | 4.08 | 0.003139 | Up |
| LOC_Os02g02560.1 | UGP2 | UTP–glucose-1-phosphate uridylyltransferase, putative | 1.25 | 0.001296 | Up |
| LOC_Os02g36924.1 | ZmMADS2 | OsMADS27 - MADS-box family gene with MIKCc type | −2.65 | 0.042858 | Down |
| LOC_Os03g07250.1 | CYP704B2 | cytochrome P450, putative, expressed | −1.04 | 9.99E-11 | Down |
| LOC_Os04g33720.1 | INV4 | glycosyl hydrolases, putative, expressed | 3.71 | 0.00555 | Up |
| LOC_Os04g57490.1 | CP1 | cysteine protease, putative, expressed | −3.77 | 0.003025 | Down |
| LOC_Os05g50380.1 | AGPL1 | glucose-1-phosphate adenylyltransferase large subunit | −2.68 | 0.035567 | Down |
| LOC_Os08g03682.1 | CYP703A3 | cytochrome P450, putative, expressed | 3.22 | 0.039267 | Up |
| LOC_Os08g43290.1 | OsC4 | LTPL44 - Protease inhibitor/seed storage/LTP family protein | 4.57 | 0.011692 | Up |
| LOC_Os09g35700.1 | LTP45 | LTPL45 - Protease inhibitor/seed storage/LTP family protein | 3.88 | 0.010751 | Up |
| LOC_Os10g26470.1 | SUT3 | sucrose transporter, putative, expressed | −2.60 | 0.040685 | Down |
| LOC_Os10g34360.1 | YY2 | stilbene synthase, putative, expressed | 4.67 | 0.002466 | Up |
| LOC_Os11g37280.1 | OsC6 | LTPL68 - Protease inhibitor/seed storage/LTP family protein | 2.52 | 0.048468 | Up |
Discussion
Spatio-temporal regulation of gene expression determines many crucial developmental changes in plants. Gene regulation may occur at transcriptional, post-transcriptional, translational and post-translational levels. In the previous report, we have shown transcriptional level regulation of bHLH142 expression. Expression of GUS reporter gene driven by bHLH142 promoter was restricted to anther tissue in transgenic rice. However, in the Arabidopsis transgenic plants, bHLH142 promoter: GUS construct showed ubiquitous expression pattern30. This suggests that bHLH142 expression is transcriptionally regulated through its promoter and anther-specific activity is restricted only to rice or monocots. In this study, we have identified other aspects related to regulation of bHLH142 expression through protein level expression. We showed that there is a temporal difference between transcript and polypeptide accumulation of bHLH142, as its polypeptide was detected at high level in anthers at tetrad and MP stage, while negligible amount of the transcript was detected in anthers at MP stage. Previously, enhanced protein accumulation in germinating pollen compared to mature pollen was observed in case of LAT52 gene in tobacco and tomato41. Delayed detection of bHLH142 polypeptide in comparison to its transcripts suggested regulation of its expression beyond transcriptional level. We hypothesise that stage-specific translation enhancement might be the reason for higher accumulation of bHLH142 protein in MP anther. Such kind of translation enhancement has previously been observed during tobacco pollen development and germination41,42. Transient as well as stable expression studies of sequence present within 5′ UTR region of LAT52 gene caused increase in translational yield in developmentally regulated manner during pollen maturation41. Similarly, 5′ UTR region of NTP303 gene also enhanced translation in pollen tube but not in mature pollen42. We found sequence similarity between 5′ UTR of bHLH142, ntp303 and LAT52 genes suggesting similar type of translation enhancement in case of bHLH142 in MP anther (Figure S6). Apart from this, the other possible reason behind biphasic protein accumulation of bHLH142 may lie in the enhancement of its protein stability. In the recent report, proteomic and phospho-proteomic analysis of rice meiotic anthers showed presence of bHLH142 protein in phospho-proteome43. Detection of bHLH142 protein in anther phospho-proteome suggested its post-translational level regulation that may change its stability. Although these hypotheses need more experimental validations to reach any specific conclusion, above-mentioned findings suggest that expression of bHLH142 is regulated at transcriptional, post-transcriptional/translational and/or post-translational levels during rice anther development.
Previous reports showed that transgenic knock-down/mutant knock-out of bHLH142 resulted in male sterile plants because of no pollen formation20,21. Mutants were defective in tapetum development and degeneration process that led to the pollen abortion. Our experiments have shown that overexpression of bHLH142 also leads to completely male sterile plants, although compromised pollen grains were formed in these plants. Ko et al.20 have shown that mutation in bHLH142 results in failure of tapetal PCD and overexpression in our study results in faster tapetal PCD. This reflects a role of bHLH142 in tapetum degeneration process. Furthermore, overexpression of bHLH142 also affected the anther dehiscence process, which caused complete male sterility. Thus, bHLH142 has other functions besides regulating tapteum degeneration during anther development in rice. The phenotype in overexpression transgenic rice is due to bHLH142 as transgenics with vector alone or transgenics made using the same vector but containing other genes have been found to be fertile44 (our unpublished work).
During the pollen development, anther wall not only forms protective covering for developing microspore but the innermost layer of the wall, tapetum also provides nutrition to them8. During the release of pollen, anther wall again plays an important role and is involved in dehiscence process. Tapetum development and its degeneration have been well studied in rice35. However, mechanism of development and degeneration of other anther wall layers is still not clear. From this study, it is evident that bHLH142 regulates development and degeneration of different anther wall layers. Poorly developed and fast degenerating tapetum was seen in developing anther, whereas lack of endothecium lignification, septum and stomium (modifications of epidermal cells) degeneration was observed in mature anther of bHLH142OE transgenic plants.
Role of bHLH142 in the development and degeneration of anther wall layers, including endothecium and modified epidermis (stomium and septum) was identified by this study. We have shown that bHLH142 polypeptide accumulated in the epidermis, pollen and vascular tissue of the mature anther and its overexpression caused defects in secondary endothecium thickening and degeneration of the septum and stomium. Lignification of endothecium appears after the vacuolated pollen stage but the genes required for the process express a little earlier45. Downregulation of the various lignin metabolism-related genes in tetrad anther might be the reason for absence of lignification in bHLH142OE anthers. Furthermore, defect in septum and stomium lysis may have occurred because of various reasons. In most of the plants, enzymatic lysis of septum occurs with the help of cell wall degrading enzymes like polygalacturonases (PGs), pectinases and expansins5,46,47. Several cell wall modification-related genes like PGs, expansins and genes encoding pectin-degrading enzymes were found to be downregulated in transgenic anthers. Furthermore, role of PCD is also suggested in septum and stomium lysis48 and downregulation of cell death-related genes in bHLH142OE MP anther may have contributed to this phenotype. Moreover, water status in anther plays crucial role in pollen development and anther dehiscence49. Role of aquaporins and sugar transporters in inducing dehydration during anther dehiscence has been reported earlier50. We detected expression of bHLH142 protein in vascular bundle of MP anther. Four aquaporin genes and one sucrose transporter gene were found to be downregulated in MP stage of transgenic anther, which suggests that defects in water conduction might have occurred that affected anther dehiscence.
Besides regulating the anther wall functions, bHLH142 also appeared to regulate pollen maturation process as most of the pollen grains of bHLH142OEplants were shrunken and non-viable. Several metabolic processes, like carbohydrate and lipid metabolism affect the pollen maturation process39. Complete maturation of pollen grains requires synthesis and transport of carbohydrate and lipids. Change in expression of various carbohydrate-related genes might have altered the synthesis and transport of sugars to the pollen grain, which resulted in their shrunken shape. Change in expression of sugar partitioning-related genes MST8, INV4, UGP2, SUT3 and AGPL140,51 suggests that defect in sugar loading/unloading may cause the formation of defective pollen in bHLH142 overexpressing plants. Lipid metabolism during the pollen maturation process is an important process as deposition of lipid is required for proper pollen wall formation. Lipid transfer proteins (LTPs) are known to be involved in pollen maturation process36,39. Change in expression of various lipid metabolism-related genes and several LTPs in bHLH142 overexpressing anther suggests that deposition of lipid on pollen wall might be altered, which contributed to the pollen abortion. CYP704B2 and CYP703A3, genes encoding cytochrome P450 enzymes, catalyse hydroxylation of fatty acid to assist the pollen wall development36,38. Both of these genes were found to have altered expression in bHLH142OE transgenic anther. Furthermore, OsC4 and OsC6, important LTPs already known for anther development, were also differentially expressed in the bHLH142OE anthers. ROS signalling has been observed to play an important role during anther cell differentiation and pollen development. Rice MADS3 has been shown to regulate ROS homeostasis during late anther development by directly regulating expression of metallothionin gene52. We detected main phenotypic difference in bHLH142OE plants during later stage of pollen development. Downregulation of metallothionin, peroxidases and glutathione-s-transferase genes in bHLH142 overexpressing anthers suggests that alteration in ROS homeostasis may have contributed to defect in pollen maturation process. Therefore, it appears that bHLH142 affects metabolic processes like carbohydrate and lipid metabolism, cell wall modification, ROS homeostasis and cell death in rice anther to control both pollen development and anther dehiscence.
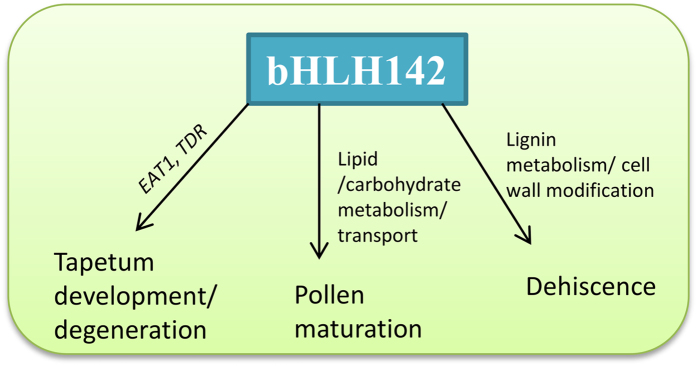
A putative model describing various functions of bHLH142 during anther development process in rice is given in Fig. 7. bHLH142 directly or indirectly affects various metabolic pathways-related genes to ensure proper anther development. It directly regulates expression of EAT1 and TDR20 required for the tapetum development and degeneration. Furthermore, it directly or indirectly affects the various lipid and carbohydrate metabolism-related genes required for pollen wall development and accumulation of starch during pollen maturation. It also affects the selective deposition of lignin and cell-wall degeneration-related genes required for the anther dehiscence. Therefore, bHLH142 acts in different stages of rice anther development to control different processes required for proper development and release of the pollen grains. Thus, this study has explored the novel functions of bHLH142 in rice anther, which will contribute to the understanding of anther development process in crops to control male fertility for the hybrid seed production.
Figure 7. Proposed model of bHLH142 functions during rice anther development.
bHLH142 regulated various aspects of anther development in rice. It directly regulates the expression of EAT1 and TDR to control tapetum development and degeneration. It affects the lipid and carbohydrate metabolism-related genes to control pollen maturation and affects lignin and cell wall modification-related genes to control anther dehiscence.
Methods
Plant material and growth conditions
For preparation of bHLH142OE transgenic plants, ORF of the gene including flanking region of 5′ UTR was amplified from rice (variety IR 64) anther cDNA using specific primer pairs (Table S1) and by using BamHI and KpnI restriction sites, fragment was cloned into plant binary vector pB4NU under maize ubiquitin promoter (Figure S2). Construct was then used for Agrobacterium-mediated rice transformation (variety Pusa Basmati1) as described by Mohanty et al.53. Transgenic plants were screened through PCR using hygromycin phosphotransferase (hpt) gene-specific primers (Table S1). WT and transgenic rice plants were grown in growth chamber (Conviron) maintained at 27 °C temperature, 70% relative humidity and 12 h dark/12 h light photoperiod with light intensity 220–350 μmol/m2s.
Phenotypic analysis
For all the phenotypic analyses, stages of WT and transgenic spikelets/anthers were characterised as mentioned by Deveshwar et al.54 and Zhang et al.1. For histological observations, WT and transgenic spikelets of different stages were fixed in FAA solution (3.7% formaldehyde, 50% ethanol, 5% glacial acetic acid) overnight at 4 °C followed by dehydration through ethanol series and embedded into Paraplast plus X-tra (Sigma-Aldrich). Paraffin sections (8–10 μm) were cut using rotary microtome (Leica Biosystems) and stained with 0.25% toluidine blue (Sigma-Aldrich) and analysed under light microscope. In figures, cross-sections of only anther parts are shown. Endothecial lignification in anthers was studied by observing WT and transgenic anther sections in UV light under florescence microscope (Nikon-80i)45. For scanning electron microscopy, post-anthesis anthers were fixed into FAA solution and treated overnight with 0.2% osmium tetroxide followed by serial ethanol dehydration. The dehydrated samples were palladium coated and examined with scanning electron microscope (Evo LS25, Zeiss)25. Pollen viability was examined by crushing WT and transgenic anthers into I2-KI solution (3:1) and pollen grains were counted under light microscope25. Images of plants and panicles were taken with Nikon-5200 DSLR camera. All the images were processed with ImageJ software (NIH, USA).
Immunoblotting and in situ immunolocalization
For immunoblotting and immunolocalization WT spikelets of different anther development stages were used. bHLH142-specific antibody was raised in rabbit against the peptide (GPFESSPTPRSGGGRKRSR). Immunoblotting and in situ immunolocalization was performed as previously described32. To check the specificity of antibody coding sequence of EAT1, TDR and bHLH142 were cloned in pET28 vector and protein was expressed in E. coli (BL21). Immunoblotting was performed using anti-bHLH142 and anti-HIS antibody against these three proteins. To check the protein level expression of bHLH142, total protein from different stages of rice spikelet were used. Total protein was extracted using protein extraction buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 50 μM PMSF, 1 mM DTT, 1 mM benzimide, and 2 μg/ml aprotenin-A). It was quantified by Bradford Reagent (Sigma-Aldrich) and equal amount of the protein from different samples was electrophoresed in 10% polyacrylamide gel and blotted onto the PVDF membranes (Mdi Membrane Technologies). Membranes were first incubated either with anti-bHLH142 (1:3000 dilutions) or anti-tubulin (Sigma-Aldrich) (1:8000 dilution) primary antibody and then with HRP-conjugated anti-rabbit secondary antibody (Sigma-Aldrich) (1:5000 dilutions). Signals were detected using the ECL Plus kit (Amersham, GE Healthcare) as per manufacturer’s instructions.
Immunolocalization was performed as described by Nayar et al.32. Briefly, rice spikelets of different stages were fixed in 4% paraformaldehyde. Paraffin sections (8 μm) of the tissue were incubated in blocking buffer (1X PBS, 5% fat-free milk) for 2 h. Sections were incubated with anti-bHLH142 primary antibody (1:100 diluted in 1X PBS, 0.1% Tween20, 3% fat-free milk) overnight and with AP-conjugated anti-rabbit secondary antibody (Sigma-Aldrich) for 2 h (1:300 dilutions). NBT-BCIP solution (Amresco Inc.) was added to the samples and kept in dark condition till appearance of visible colour. Slides were mounted into DPX mounting medium (Sigma-Aldrich) and photographed under NIKON 80i microscope.
qRT-PCR analysis
Total RNA was isolated using TRI-Reagent (Sigma-Aldrich) and purity and concentration of the RNA samples were checked by NANODROP 2000 Spectrophotometer (Thermo Scientific). cDNA for different samples were prepared with 1 μg of RNA using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). qRT-PCR analysis was performed with three biological and three technical replicates of each sample using ABI 7500 fast qRT-PCR system with the ‘Fast SYBR Green master mix’ (Applied Biosystems). Specific gene expression was normalised to ubiquitin5 (UBQ5) as internal control gene.
TUNEL assay
TUNEL assay was performed as previously described14. Briefly, FAA-fixed tissues were used to perform TUNEL assay. Paraffin sections of WT and transgenic rice spikelets were prepared as described earlier. Sections were deparaffinised and treated with 20 mg/ml proteinase K enzyme in humid chamber for 10 min followed by PBS wash. After brief fixation of 10 min with 4% (w/v) paraformaldehyde and PBS wash, nick-end labelling of the fragmented DNA was performed using Dead End Fluorometric TUNEL system (Promega) kit according to the manufacturer’s instructions. Samples were counter stained with propidium iodide and analysed under confocal laser scanning microscope (Leica TCS SP5) with 488-nm/510-nm excitation/emission spectrum for fluorescein and a 530-nm/640-nm excitation/emission spectrum for propidium iodide.
Subcellular localization
Nuclear localization signal (NLS) in bHLH142 protein sequence was predicted by cNLS Mapper programme. bHLH142 CDS (Coding DNA Sequence) was fused to C-terminal sequence of YFP by cloning into pSITE3CA vector using gateway technology (Invitrogen). Empty vector and fusion constructs were transiently expressed in onion epidermal cells through biolistic bombardment-based transformation as described previously55. YFP fluorescence was analysed under confocal microscope (Leica TCS SP2).
RNA-Seq analysis
Total RNA was extracted from tetrad and MP stage of WT and transgenic anthers using Tri-Reagent (Sigma-Aldrich) and purified using RNeasy MinElute CleanUp Kit (Qiagen) after DNAse I treatment. Quality and integrity of RNA was checked by NANODROP 2000 Spectrophotometer (Thermo Scientific) and Bioanalyser 2100 (Agilent Technologies). RNA sample of each genotype (WT and bHLH142OE) were sequenced on Illumina HiSeq platform as per manufacturer instructions. More than 60 million raw reads giving more than 12 Gb sequence data for each sample were obtained by paired-end sequencing. High quality reads were filtered and then aligned to rice gene model downloaded from RGAP ver. 7.0. Uniquely mapped reads were used for expression analysis. DESeq program56 was used to perform differential gene expression analysis. Count data given as input in DESeq programme were obtained using HTSeq of Python package from the alignment files. Genes showing log2 fold change ≥1 and p-value ≤0.05 compared to WT were treated as differentially expressed genes. GO enrichment was performed using BiNGO tool57 and map was visualised in Cytoscape programme. Putative function of each differentially expressed gene was obtained from RGAP Ver. 7.0.
Additional Information
How to cite this article: Ranjan, R. et al. bHLH142 regulates various metabolic pathway-related genes to affect pollen development and anther dehiscence in rice. Sci. Rep. 7, 43397; doi: 10.1038/srep43397 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work is supported by grant from NIPGR, New Delhi and project grant from the DBT (Department of Biotechnology), Government of India. R.R. and N.M. acknowledge CSIR (Council of Scientific and Industrial Research) for Junior and Senior Research Fellowship awards. R.K. acknowledges UGC (University Grant Commission) for Dr. D.S. Kothari Post-Doctoral Fellowship award.
Footnotes
The authors declare no competing financial interests.
Author Contributions R.R. conducted all the experiments, analysed the data and drafted the manuscript. R.K. performed subcellular localization study. N.M. was involved in phenotypic studies and RNA-seq data analysis and S.B. participated in crossing experiments. A.K.T. conceived, designed and supervised the study, and along with S.K.P. and S.K. analysed all data and guided in the manuscript preparation. All authors have contributed in manuscript writing and approved the final version.
References
- Zhang D., Luo X. & Zhu L. Cytological analysis and genetic control of rice anther development. J. Genet. Genomics 38, 379–390 (2011). [DOI] [PubMed] [Google Scholar]
- Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434 (2005). [DOI] [PubMed] [Google Scholar]
- Walbot V. & Egger R. L. Pre-meiotic anther development: cell fate specification and differentiation. Annu. Rev. Plant Biol. 67, 365–395 (2016). [DOI] [PubMed] [Google Scholar]
- Matsui T., Omasa K. & Horie T. Mechanism of anther dehiscence in rice (Oryza sativa L.). Ann. Bot. 84, 501–506 (1999). [Google Scholar]
- Wilson Z. A., Song J., Taylor B. & Yang C. The final split: the regulation of anther dehiscence. J. Exp. Bot. 62, 1633–1649 (2011). [DOI] [PubMed] [Google Scholar]
- Wu H. M. & Cheun A. Y. Programmed cell death in plant reproduction. Plant Mol. Biol. 44, 267–281 (2000). [DOI] [PubMed] [Google Scholar]
- Scott R. J., Spielman M. & Dickinson H. G. Stamen structure and function. Plant Cell 16 Suppl, S46–60 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R. B., Beals T. P. & Sanders P. M. Anther development: basic principles and practical applications. Plant Cell 5, 1217–1229 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana R., Kapoor S. & Tyagi A. K. Anthology of anther/pollen-specific promoters and transcription factors. Crit. Rev. Plant Sci. 31, 359–390 (2012). [Google Scholar]
- Zhang W. et al. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133, 3085–3095 (2006). [DOI] [PubMed] [Google Scholar]
- Jung K. H. et al. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17, 2705–2722 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. et al. Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. 72, 612–624 (2012). [DOI] [PubMed] [Google Scholar]
- Zhu E. et al. The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 83, 976–990 (2015). [DOI] [PubMed] [Google Scholar]
- Li N. et al. The rice Tapetum Degeneration Retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999–3014 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen A. M. et al. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33, 413–423 (2003). [DOI] [PubMed] [Google Scholar]
- Xu J. et al. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22, 91–107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Feng B. & Ma H. AMS-dependent and independent regulation of anther transcriptome and comparison with those affected by other Arabidopsis anther genes. BMC Plant Biol. 12, 23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. S. et al. Tapetum Degeneration Retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 1, 599–610 (2008). [DOI] [PubMed] [Google Scholar]
- Niu N. et al. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun. 4, 1445 (2013). [DOI] [PubMed] [Google Scholar]
- Ko S. S. et al. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 26, 2486–2504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. et al. The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26, 1512–1524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzer C. J. The processes of anther dehiscence and pollen dispersal. I. The opening mechanism of longitudinally dehiscing anthers. New Phytol. 105, 487–489 (1987). [DOI] [PubMed] [Google Scholar]
- Wilson Z. A. & Zhang D. B. From Arabidopsis to rice: pathways in pollen development. J. Exp. Bot. 60, 1479–1492 (2009). [DOI] [PubMed] [Google Scholar]
- Thangasamy S. et al. Rice SIZ1, a SUMO E3 ligase, controls spikelet fertility through regulation of anther dehiscence. New Phytol. 189, 869–882 (2011). [DOI] [PubMed] [Google Scholar]
- Zhou S. et al. Pollen Semi-Sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23, 111–129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q. H., Ramm K., Shivakkumar R., Dennis E. S. & Upadhyaya N. M. The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol. 135, 1514–1525 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. et al. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19, 534–548 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Lange S. et al. Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 34, 519–528 (2003). [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Seki M., Shinozaki K. & Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana R., Kapoor S. & Tyagi A. K. Spatial and temporal activity of upstream regulatory regions of rice anther-specific genes in transgenic rice and Arabidopsis. Transgenic Res. 22, 31–46 (2013). [DOI] [PubMed] [Google Scholar]
- Yang S. L. et al. Overexpression of TAPETUM DETERMINANT1 alters the cell fates in the Arabidopsis carpel and tapetum via genetic interaction with excess microsporocytes1/extra sporogenous cells. Plant Physiol. 139, 186–191 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayar S., Sharma R., Tyagi A. K. & Kapoor S. Functional delineation of rice MADS29 reveals its role in embryo and endosperm development by affecting hormone homeostasis. J. Exp. Bot. 64, 4239–4253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray F., Kalla R., Jacobsen J. & Gubler F. A role for HvGAMYB in anther development. Plant J. 33, 481–491 (2003). [DOI] [PubMed] [Google Scholar]
- Huang J. et al. Ectopic expression of TAPETUM DETERMINANT1 affects ovule development in Arabidopsis. J. Exp. Bot. 67, 1311–1326 (2016). [DOI] [PubMed] [Google Scholar]
- Zhang D. & Yang L. Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 17, 49–55 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang D. et al. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154, 149–162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. Cytochrome P450 family member CYP704B2 catalyzes the {omega}-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22, 173–190 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. et al. Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integr. Plant Biol. 56, 979–994 (2014). [DOI] [PubMed] [Google Scholar]
- Shi J., Cui M., Yang L., Kim Y. J. & Zhang D. Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 20, 741–753 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang H. et al. Carbon Starved Anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22, 672–689 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate N., Spurr C., Foster G. D. & Twell D. Maturation-specific translational enhancement mediated by the 5′-UTR of a late pollen transcript. Plant J. 10, 613–623 (1996). [DOI] [PubMed] [Google Scholar]
- Hulzink R. J. et al. The 5′-untranslated region of the ntp303 gene strongly enhances translation during pollen tube growth, but not during pollen maturation. Plant Physiol. 129, 342–353 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. et al. Proteomic and phosphoproteomic analyses reveal extensive phosphorylation of regulatory proteins in developing rice anthers. Plant J. 84, 527–544 (2015). [DOI] [PubMed] [Google Scholar]
- Dansana P. K., Kothari K. S., Vij S. & Tyagi A. K. OsiSAP1 overexpression improves water-deficit stress tolerance in transgenic rice by affecting expression of endogenous stress-related genes. Plant Cell Rep. 33, 1425–1440 (2014). [DOI] [PubMed] [Google Scholar]
- Cecchetti V., Altamura M. M., Falasca G., Costantino P. & Cardarelli M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20, 1760–1774 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorguet B., Schipper D., van Lammeren A., Visser R. G. & van Heusden A. W. ps-2, the gene responsible for functional sterility in tomato, due to non-dehiscent anthers, is the result of a mutation in a novel polygalacturonase gene. Theor. Appl. Genet. 118, 1199–1209 (2009). [DOI] [PubMed] [Google Scholar]
- Ogawa M., Kay P., Wilson S. & Swain S. M. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatore A., Trobacher C. P. & Greenwood J. S. Ricinosomes predict programmed cell death leading to anther dehiscence in tomato. Plant Physiol. 149, 775–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firon N., Nepi M. & Pacini E. Water status and associated processes mark critical stages in pollen development and functioning. Ann. Bot. 109, 1201–1214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bots M. et al. Aquaporins of the PIP2 class are required for efficient anther dehiscence in tobacco. Plant Physiol. 137, 1049–1056 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. et al. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J. 82, 570–581 (2015). [DOI] [PubMed] [Google Scholar]
- Hu L. et al. Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23, 515–533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty A., Sarma N. P. & Tyagi A. K. Agrobacterium-mediated high frequency transformation of an elite indica rice variety Pusa Basmati 1 and transmission of the transgenes to R2 progeny. Plant Sci. 147, 127–137 (1999). [Google Scholar]
- Deveshwar P., Bovill W. D., Sharma R., Able J. A. & Kapoor S. Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biol. 11, 78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G., Giri J. & Tyagi A. K. Rice OsiSAP7 negatively regulates ABA stress signalling and imparts sensitivity to water-deficit stress in Arabidopsis. Plant Sci. 237, 80–92 (2015). [DOI] [PubMed] [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K. & Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.