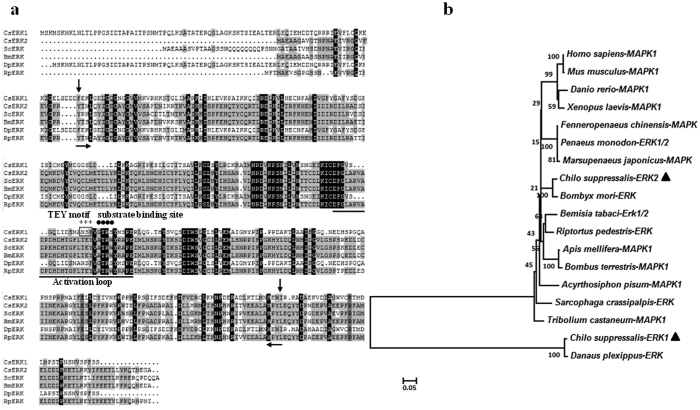
Figure 3. Comparison of C. suppressalis ERK1 and ERK2 MAPK genes (CsERK1 and CsERK2) with known ERK MAPKs from other species.
(a) Amino acids with 100%, 75%, and 50% conservation are shaded in black, dark grey and light grey. The predicted S_TKc domain was found at position 90–369 (arrow) of CsERK1 and 28–316 (arrow) of ERK2. The characteristic ERK structure, activation loop (underline), substrate binding site (dark spot), and TEY motif, are indicated by asterisks. (b) Phylogenetic tree of relationships between CsERKs and MAPKs from other species (Supplementary Table S2). The bar indicates a phylogenetic distance of 0.05, the position of CsERK is indicated by a black triangle.