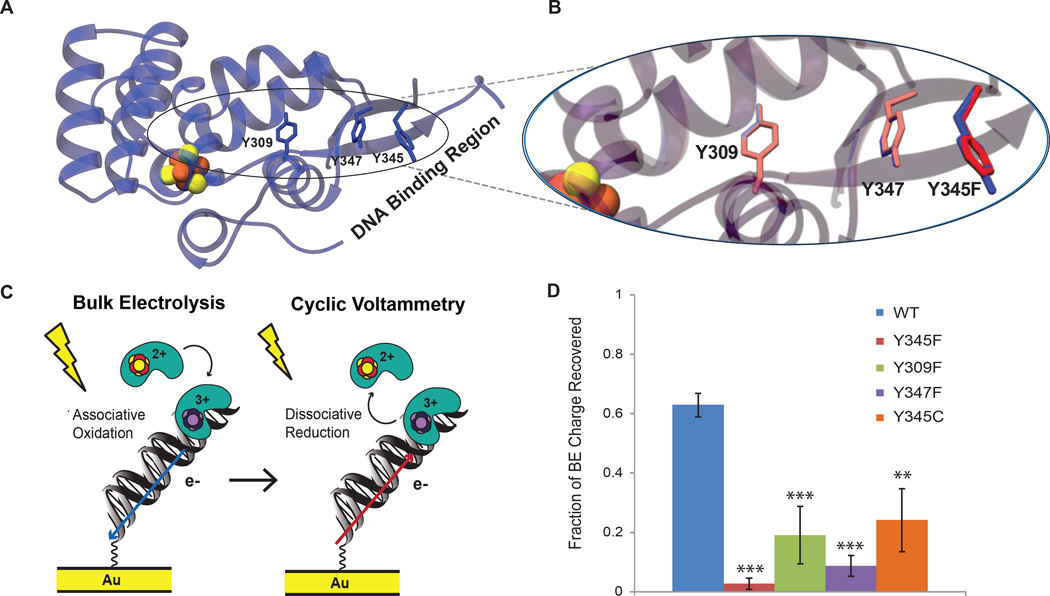
Figure 3. DNA-binding, charge-transfer deficient p58C mutants.
A) Tyrosine residues conserved in eukaryotic primase [4Fe4S] domain (Y309, Y345, Y347 in H. sapiens, blue sticks) are located between the [4Fe4S] cluster (orange and yellow spheres) and DNA-binding region. The DNA binding region, consisting of residues R302, R306, K314, and W327, is located ~20–30 Å from the cluster, necessitating electron transfer through the protein matrix for exchange of charge between the [4Fe4S] cofactor and bound DNA. B) Expanded region of the overlaid crystal structures of p58C (PDB 3L9Q, blue) and p58C Y345F (PDB 517M, red) demonstrates the minimal structural impact of the Y-F mutation; the phenylalanine residue in the mutant adapts the same orientation as the tyrosine in WT p58C. All mutants bind DNA with approximately the same affinity as WT p58C. C) Scheme depicts redox reactions in electrochemical assays with wild type and mutant p58C. Bulk electrolysis first oxidizes p58C and promotes tight DNA binding. CV then reduces the DNA-bound protein, converting it to the weakly associated, electrochemically inactive form. Both require the tyrosine charge transfer pathway and must be accounted for when comparing charge transfer proficiency. D) WT p58C recovers significantly more (63±4%) bulk electrolysis charge than the mutants, suggesting that perturbation of the charge transfer pathway diminishes DNA-bound redox chemistry and consequently affects the redox switch. All bulk electrolysis reactions and CV scans were performed on 16 µM p58C/mutant in 20 mM Tris, pH 7.2, 75 mM NaCl at a 100 mV/s scan rate for CV, using a Ag/AgCl reference electrode. Mean ± SD of n=3 scans per variant, ** = 0.001<p<0.0005, *** = p<0.0005, student’s t-test.