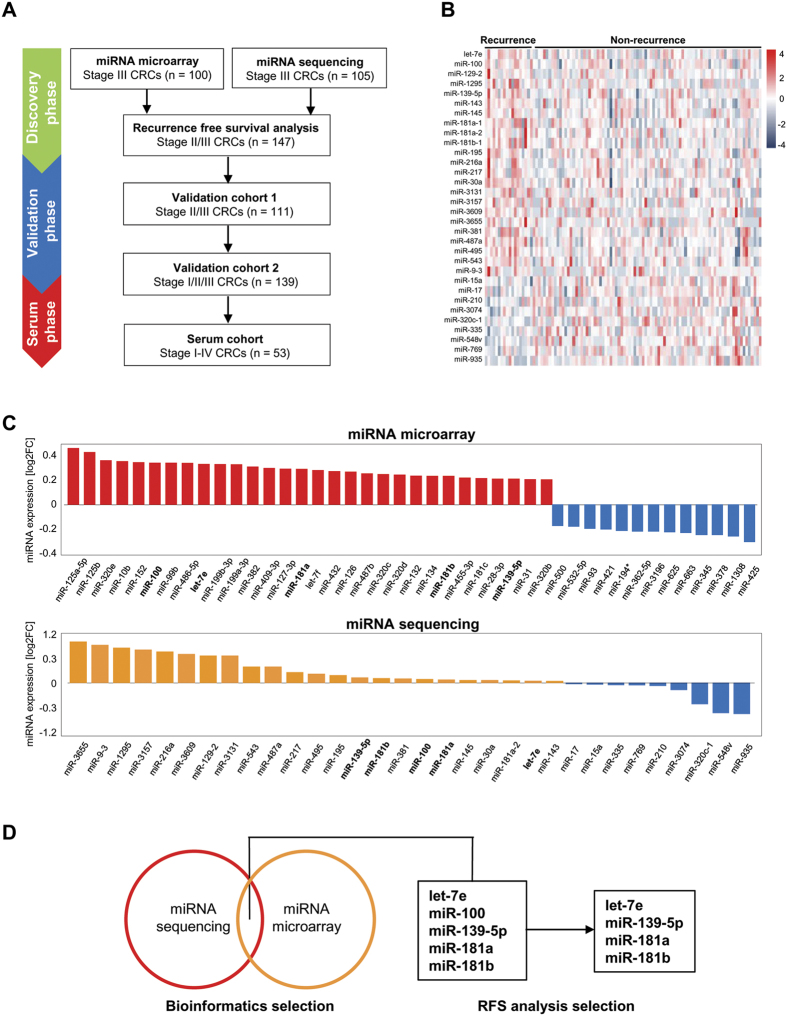
Figure 1. Selection of candidate miRNAs by miRNA sequencing and microarray analysis.
(A) Strategy for the identification of CRC recurrence-specific miRNAs. (B) Heat map of miRNA sequencing expression by the TCGA data. (C) Ranking of miRNAs which were significantly differentially expressed between recurrence positive and negative stage III patients by miRNA microarray in Mayo Clinic cohort and by miRNA sequencing in TCGA dataset cohort. (D) Five candidate miRNAs were significantly up-regulated in recurrence-positive CRC patients in both datasets. And 4 out of 5 miRNAs could differentiate CRC patients who experienced recurrence versus those who did not, by measuring RFS using Kaplan-Meier analysis in stage II/III CRC patients from the TCGA cohort.