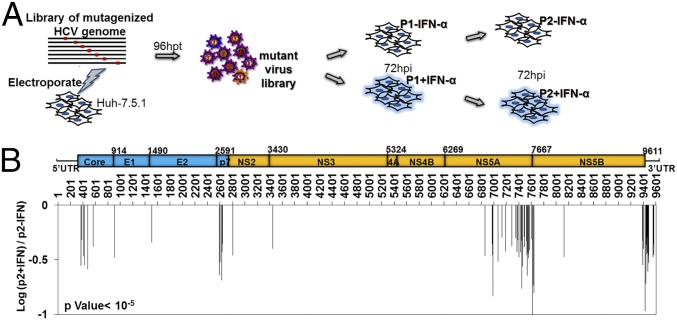
Fig. 1.
Genomic screen of mutant virus library with IFN-α treatment. (A) Schematic diagram of selection to identify viral sequences critical for counteracting IFN-α responses. A 15-nt insertional mutant HCV library was subjected to infection with Huh-7.5.1 cells in the presence or absence of IFN-α treatment at the IC50 concentration (1 U/mL; Fig. S1) for two rounds, and the supernatant was collected (p2). (B) IFN-sensitive mutations are clustered at four regions on the virus genome: the N terminus of core, the N terminus of p7, NS5A domains II and III, and the 3′UTR. The x axis indicates the positions of the 15-nt insertion on the genome. The y axis shows the ratio of mutant frequency with IFN-α treatment to mutant frequency without IFN-α treatment. The schematic picture above the histogram shows the FNX-HCV virus genome composition. Blue is from the J6 strain, and yellow is from the JFH1 strain.