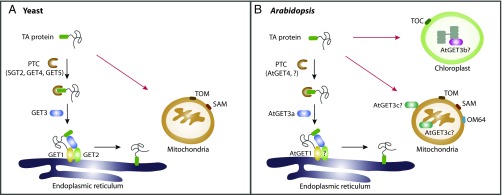
Fig. 1.
Working model for the GET system. (A) In yeast, some TA proteins targeted to the ER membrane are GET dependent (black arrows), whereas targeting to mitochondria is GET independent and uses receptors including TOM and SAM complexes (red arrows). In the GET pathway, the newly synthesized protein is initially recognized by the PTC complex (SGT2 and GET4–GET5), and then is transferred to the ATPase GET3. Subsequently, cytosolic GET3 guides the TA protein to the ER membrane via interacting with the ER-localized GET1–GET2 complex, and finally promotes the insertion of the TA protein into the ER membrane. (B) In Arabidopsis, the AtGET3a-dependent pathway is involved in TA protein insertion into the ER (black arrows). TA protein transport to mitochondria and chloroplast is likely GET independent (red arrows) and requires distinct receptors, including TOM and SAM complexes, as well as a plant-specific receptor OM64 for mitochondria and TOC complex for chloroplast. For TA proteins targeted to the ER, the PTC complex (AtGET4 and possibly other unidentified components?) binds to the nascent proteins and recruits the cytosolic AtGET3a. Then, AtGET3a interacts with AtGET1 and a possible coreceptor (?) to transfer the TA protein to AtGET1 for insertion into the ER. In contrast, AtGET3b is distributed in chloroplast stroma, whereas AtGET3c is found in the mitochondrial matrix or on the mitochondrial outer membrane. The biological function of both AtGET3b and AtGET3c in chloroplast or mitochondrial remains elusive.