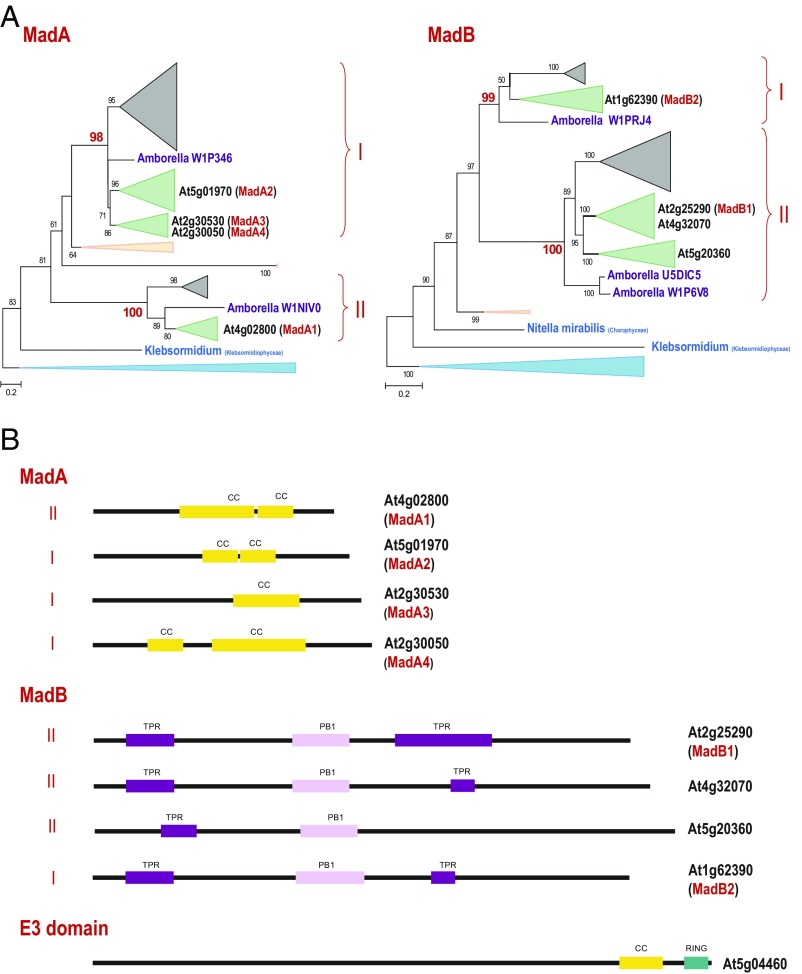
Fig. 6.
Phylogenies of the MadA and MadB families and sequence features of the plant myosin interactors. (A) Maximum-likelihood unrooted trees of the MadA and MadB families. Bootstrap values (percentage points) are shown for all internal branches. The key bootstrap values supporting subfamilies are shown in red. The collapsed branches are shown by triangles according to the respective plant taxa, as follows: moss and spikemoss in light peach; monocots in gray; dicots in green; Chlorophyta in light blue; Amborella, a basal Magnoliophyta plant, in purple; and Klebsormidium, a filamentous terrestrial alga, in teal blue. For A. thaliana, the gene identifiers are highlighted (bold). The myosin-interacting proteins are shown in red in parentheses. The complete trees in the Newick format are provided in SI Appendix as Datasets S5 (MadA) and S6 (MadB). (B) Sequence features of the putative myosin adaptor families. The lengths of the proteins and identified domains, presented as boxes, are drawn roughly to scale. CC, coiled-coil domain; PR1, PR1 domain; RING, RING finger (E3-like domain); TPR, TPR repeats.