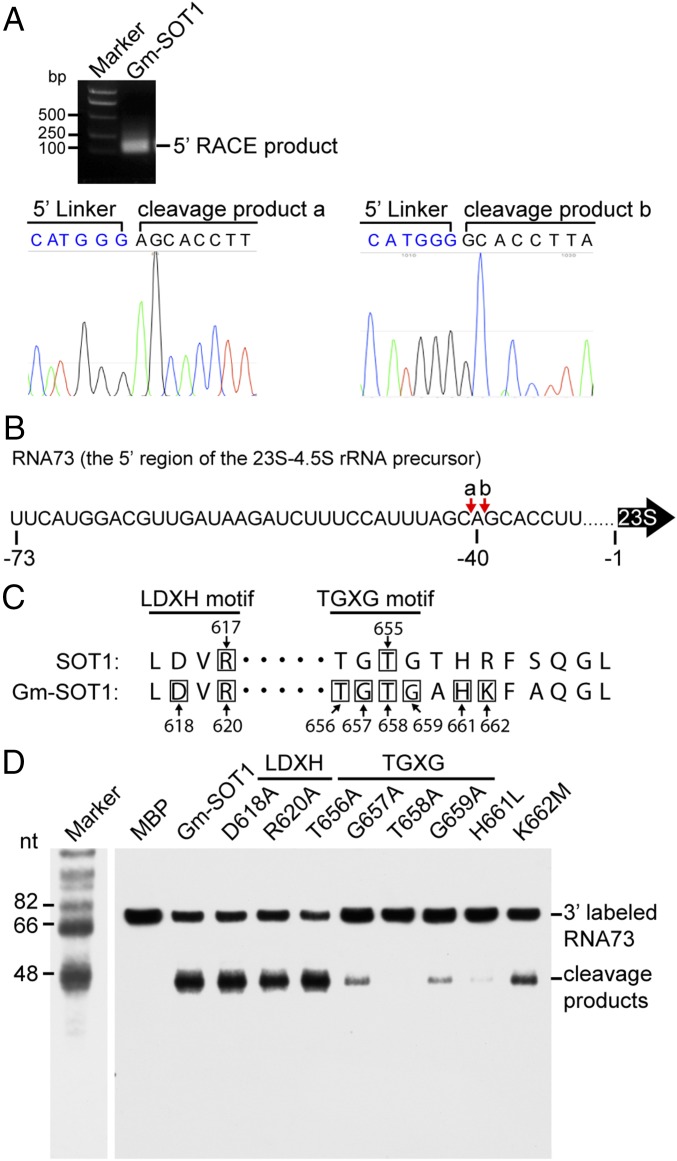
Fig. 4.
Cleavage sites in RNA73 by Gm-SOT1 and critical amino acids in the SMR domain of SOT1 for RNA endonuclease activity. (A) Mapping of the sites in RNA73 cleaved by Gm-SOT1. The cleavage products in RNA73 produced by Gm-SOT1 were recovered from the SDS/PAGE gel and reverse transcribed to cDNA by 5′ RACE, followed by sequencing. The sequencing chromatograms were constructed using four peaks (nucleobytes.com/4peaks/). (B) Schematic illustration of the cleavage sites in RNA73. The cleavage sites are indicated by red arrows and shown above the sequence of RNA73. The numbers below the sequence indicate the positions from the 5′ end of mature 23S rRNA. (C) Alignment of conserved LDXH and TGXG motifs of the SMR domain of SOT1 and Gm-SOT1. The numbers indicate the amino acid positions with respect to the translation initiation site. Black boxes indicate the amino acids in site-directed mutagenesis. (D) The TGXG motif is critical for the RNA endonuclease activity of Gm-SOT1. Site-directed mutagenesis was performed to investigate the effects of amino acids in the LDXH motif and TGXG motifs on the RNA endonuclease activity of Gm-SOT1. A total of 10 nM 3′-end biotin-labeled RNA73 was incubated with 100 nM MBP, Gm-SOT1, and Gm-SOT1 variants at 25 °C for 30 min, followed by separation of RNA on 10% (wt/vol) denaturing polyacrylamide gels. Marker sizes are shown at Left.