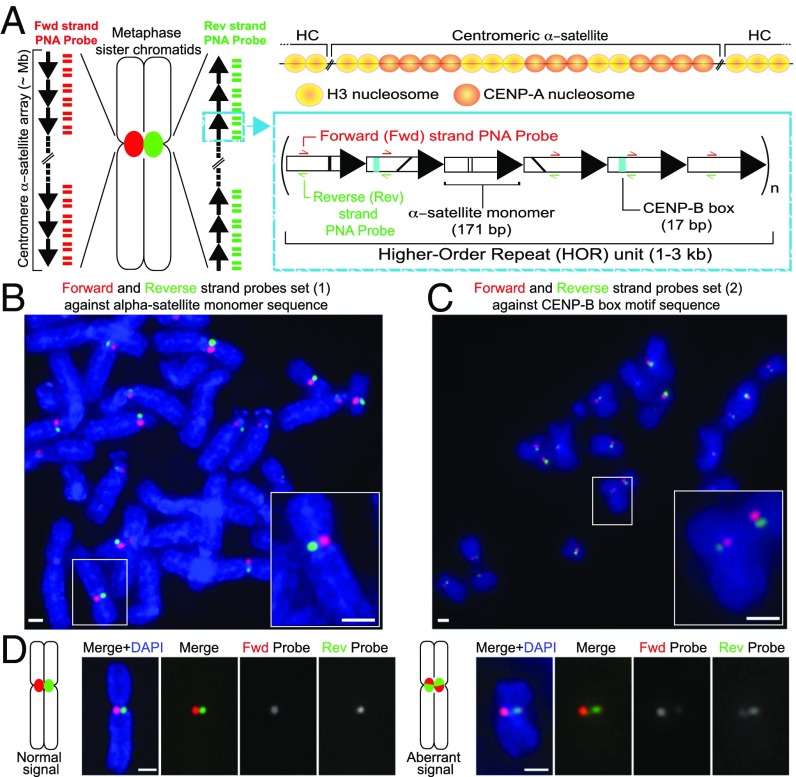
Fig. 1.
CO-FISH application to monitor human centromeric α-satellite repeats. (A) Diagram of the human centromere. (Left) The centromere locus of metaphase sister chromatids is depicted in red or green due to hybridization by unidirectional PNA probes, differentially labeled to detect the forward or reverse strands of one sister chromatid. Each black arrow symbolizes a HOR in the α-satellite array. The blue box gives a zoom in into a schematic example of the HOR tandem repeat monomers arranged head to tail. Represented on each monomer are black lines indicating divergence of sequence, teal box where the CENP-B box sequence is found, and the regions where the probes hybridize. (Above) The localization of CENP-A nucleosomes shown at the core centromere repeats, interspersed with canonical H3 nucleosomes. Outside of the core domain into the pericentromeric heterochromatin (HC), only H3 nucleosomes are found. (B) Unidirectional PNA probe set 1 against α-satellite or (C) PNA probe set 2 against the CENP-B box. White box indicates zoomed-in Insets. (D) Diagrammatic figures and example images of c-CO-FISH using probe set 1 in hTERT-RPE1 cells showing separated signal, where only one probe hybridizes on each sister chromatid (normal) and aberrant c-CO-FISH pattern, where the signal has exchanged between the two sister chromatids and probes overlap. (All scale bars, 1 μm.)