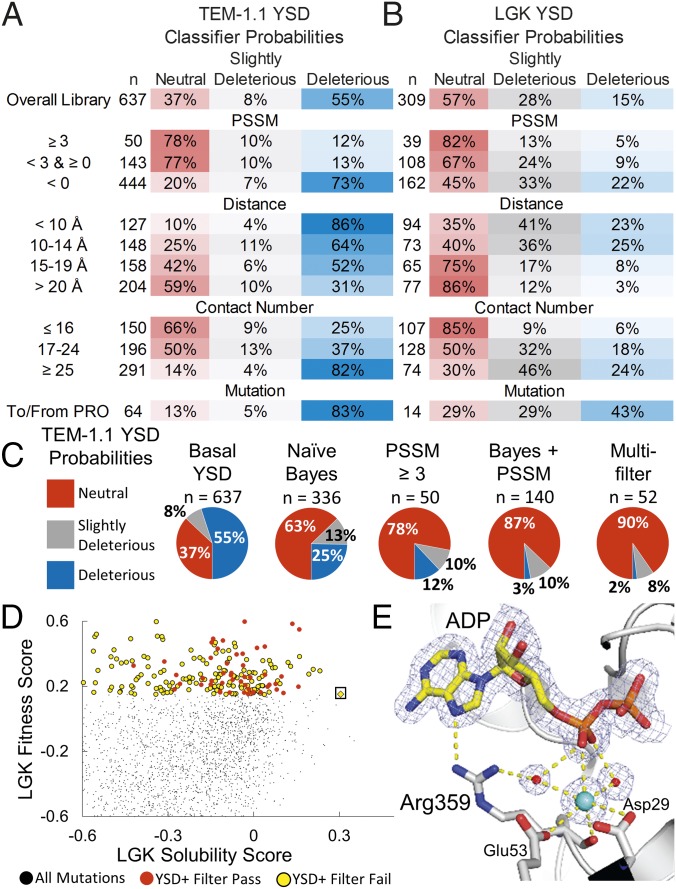
Fig. 4.
Classification methods improve probabilities of selecting mutations conferring solubility and activity but remove rare, globally optimal mutations. Classifier probabilities for YSD deep mutational scan for TEM-1.1 (A) and LGK (B). The total number of mutations found in a given bin (n) is provided, and the PSSM represents the site-specific preferences found in the evolutionary history of the enzyme. (C) Classification methods improve probabilities of selecting neutral mutations. (D) LGK fitness versus the LGK solubility score of individual mutations. Beneficial mutations from the YSD screen are shown as circles colored by whether they pass (red) or fail (yellow) the multiple-filter classification method. The Pareto optimal mutation G359R (boxed) fails the filtering due to its close distance to the active site, low evolutionary conservation, and high contact number. (E) Crystal structure of LGK G359R (PDB ID code 5TKR). G359R makes direct and water-mediated hydrogen bonds with ADP near the active site. A potassium ion also appears to be coordinated in this region, possibly contributing to the stability of the enzyme. Carbon atoms are shown in gray and yellow for the protein and ligand atoms, respectively. Nitrogen, oxygen, and phosphorous atoms are shown in blue, red, and orange, respectively. Waters and the potassium are shown as red and cyan spheres, respectively. The 2mFo-DFc electron density map is contoured to 1σ. For clarity, the magnesiums in the active site have been omitted from the figure.