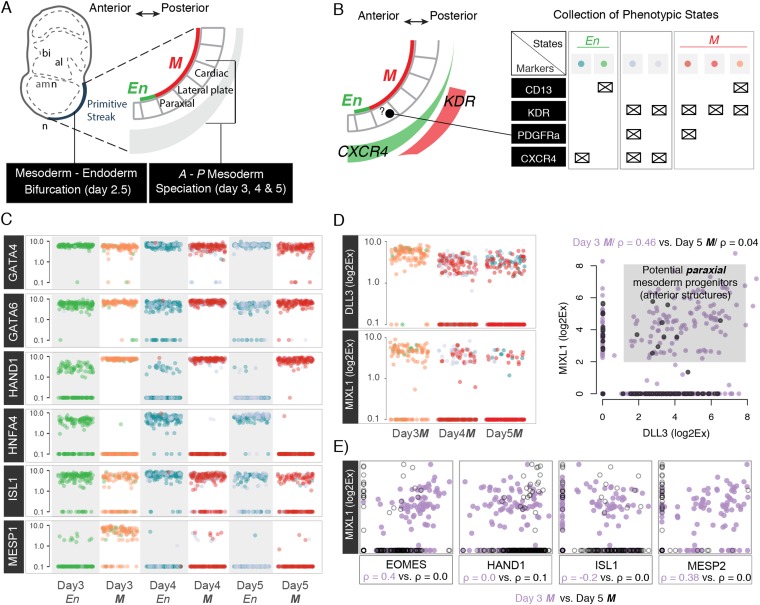
Fig. S7.
Cell states during anterior–posterior mesoderm specification. (A) Model of the anterior–posterior patterning of PS. (B) Distribution of cell surface molecules in PS and collected phenotypic states that are related to En or M cell fates. For instance, KDR+ cells are primarily related to lateral mesoderm fate, whereas PDGFRa is mainly a paraxial mesoderm marker. The color scheme is used below to indicate how different transcription factors are distributed between each phenotype. (C) Plots of Log2Ex values for six transcriptions factors that define M or En cell fates: GATA4 and GATA6 indicate both cell fates; HAND1, ISL1, and MESP1 indicate cardiac cell fates/midposterior PS; and HNFA4 indicates En cell fates/anterior PS. (D) MIXL1 (a transcription factor) and DLL3 (a Notch ligand) have been recently reported (4) as paraxial mesoderm markers. They can be detected between days 3 and 5, but they correlate only during day 3 (correlation plot), suggesting that there might be a paraxial mesoderm population early during mesoderm specification process. (E) Correlation plots for days 3 and 5 gene expression between MIXL1 (a paraxial mesoderm regulator) and four other transcription factors. Notice that EOMES that is associated with anterior PS specification has a positive correlation with MIXL1, whereas ISL1 that is expressed in the most posterior parts is slightly anticorrelated with MIXL1. However, all correlations are lost by day 5.