Significance
At this time, multidrug-resistant gram-negative bacteria are estimated to cause approximately 700,000 deaths per year globally, with a prediction that this figure could reach 10 million a year by 2050. Antivirulence therapy, in which virulence mechanisms of a pathogen are chemically inactivated, represents a promising approach to the development of treatment options. The family of lipid A phosphoethanolamine transferases in gram-negative bacteria confers bacterial resistance to innate immune defensins and colistin antibiotics. The development of inhibitors to block lipid A phosphoethanolamine transferase could improve innate immune clearance and extend the usefulness of colistin antibiotics. The solved crystal structure and biophysical studies suggest that the enzyme undergoes large conformational changes to enable binding and catalysis of two very differently sized substrates.
Keywords: lipid modification, multidrug resistance, molecular dynamics, Neisseria, membrane protein structure
Abstract
Multidrug-resistant (MDR) gram-negative bacteria have increased the prevalence of fatal sepsis in modern times. Colistin is a cationic antimicrobial peptide (CAMP) antibiotic that permeabilizes the bacterial outer membrane (OM) and has been used to treat these infections. The OM outer leaflet is comprised of endotoxin containing lipid A, which can be modified to increase resistance to CAMPs and prevent clearance by the innate immune response. One type of lipid A modification involves the addition of phosphoethanolamine to the 1 and 4′ headgroup positions by phosphoethanolamine transferases. Previous structural work on a truncated form of this enzyme suggested that the full-length protein was required for correct lipid substrate binding and catalysis. We now report the crystal structure of a full-length lipid A phosphoethanolamine transferase from Neisseria meningitidis, determined to 2.75-Å resolution. The structure reveals a previously uncharacterized helical membrane domain and a periplasmic facing soluble domain. The domains are linked by a helix that runs along the membrane surface interacting with the phospholipid head groups. Two helices located in a periplasmic loop between two transmembrane helices contain conserved charged residues and are implicated in substrate binding. Intrinsic fluorescence, limited proteolysis, and molecular dynamics studies suggest the protein may sample different conformational states to enable the binding of two very different- sized lipid substrates. These results provide insights into the mechanism of endotoxin modification and will aid a structure-guided rational drug design approach to treating multidrug-resistant bacterial infections.
Recent reports by the World Health Organization and the US Centers for Disease Control and Prevention highlight the serious and growing public health threat and high mortality rate associated with multidrug-resistant (MDR) gram-negative bacterial infections (1, 2). Increasingly, colistin is used for the treatment of sepsis caused by carbapenemase-producing gram-negative bacteria. However, this treatment has resulted in the selection of MDR bacterial isolates with chromosomal resistance mutations in lipid A biosynthesis and regulation (3) and, more recently, the appearance of a mobile colistin resistance gene (mcr-1) (4–6). Mcr-1 encodes a member of the family of phosphoethanolamine (PEA) transferases that decorates the lipid A headgroups of lipopolysaccharide with PEA. Modification of lipid A on the 1 and 4′ headgroup positions with PEA or 4-amino-arabinose masks the negatively charged phosphate groups on the bacterial surface, which are involved in interaction with cationic antimicrobial peptides (CAMPs) such as colistin and polymyxin B (7). This modification confers resistance to CAMPs, as well as host innate immune defensins; however, the exact mechanism of resistance is not known.
PEA transferases belong to the alkaline phosphatase superfamily. Many MDR gram-negative bacteria possess multiple members of this family of enzymes that are engaged in the decoration of lipid A or the conserved inner core of the lipopolysaccharide (8, 9). Neisseria gonorrheae (Ng) and Neisseria meningitidis (Nm) are closely related obligate human pathogens that are intrinsically resistant to colistin and decorate lipid A exclusively with PEA, using the enzyme lipid A PEA transferase A (EC 2.7.4.30) [EptA; lipid A PEA transferase A (LptA) in Nm was originally named by Cox et al. (10); however, LptA is also the nomenclature used for the lipopolysaccharide transport system in Escherichia coli (11). To alleviate the confusion in terminology and maintain consistency with the published nomenclature used for the enzyme from other organisms, the enzyme from Nm has been redefined as NmEptA. An update of the nomenclature for all Neisserial lipid A PEA transferase enzymes will be announced on PubMLST (pubmlst.org/)]. Mutants of NmEptA increase bacterial sensitivity to CAMPs (12), resulting in attenuation in mice and human models of infection (13). EptA catalyzes the transfer of PEA from phosphatidylethanolamine (PE), via a PEA-enzyme intermediate, to the 1 and 4′ headgroups of the lipid A of lipooligosaccharide (SI Appendix, Scheme S1). MDR-Ng has recently been identified as an urgent public health threat because of the increased burden of the sexually transmitted disease gonorrhea and the increased risk for sequelae, which includes infertility. Because of the essential involvement of EptA in the establishment of gonorrhea, EptA has been identified as a potential target for the rational design of enzyme inhibitors as therapeutic agents to treat MDR-Ng and gram-negative bacteria harboring mcr-1.
The structures of the soluble periplasmic domain of EptA from Nm (14), and the homologs, EptC, from Campylobacter jejuni (15) and Mcr-1 in E. coli (16), all of which add PEA to lipid A, have been determined (17–20). These structures all reveal a hydrolase fold similar to that of phosphonate monoester hydrolase (21) and arylsulfatase (22–24). A bound Zn2+ ion is tetrahedrally coordinated by conserved residues (His453, Asp452, Glu240, and Thr280 in NmEptA). Thr280 is the catalytic nucleophile required for PEA transfer and is present in the NmEptA structure as a phosphothreonine. Substitution of Thr280 with alanine in Mcr-1 decreases the inhibitory concentration of colistin and polymyxin B, supporting its importance in catalysis (16). On the basis of the truncated structure, it was suggested that the membrane domain would be required for the correct binding and orientation of the lipid substrates, PE and lipid A (14). To further understand the molecular details involved in substrate binding and enzyme catalysis, we have determined the crystal structure of the full-length form of NmEptA and have carried out biophysical studies aimed at assessing the ability of the enzyme to accommodate different substrates.
Results and Discussion
Purification of Full-Length NmEptA.
Recombinant NmEptA was expressed and was purified in the presence of the detergent dodecyl-β-d-maltoside (DDM). To assess the activity of the recombinant enzyme in DDM micelles, two types of enzymatic assays were performed. A fluorescence-based TLC enzyme assay was carried out using a substrate tagged with a fluorescent label, 1-acyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphoethanolamine (NBD-PEA) (SI Appendix, Fig. S2). The product formed by reaction with the enzyme was confirmed by mass spectrometry to be 1-acyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycerol. This showed that the full-length NmEptA in DDM was able to successfully remove PEA from the lipid substrate. In contrast, a soluble domain-only construct was unable to catalyze PEA hydrolysis, confirming that the transmembrane domain of NmEptA is required for its activity on a lipid substrate.
In addition, the PEA transfer from PE to lipid A of N. flavescens by recombinant NmEptA solubilized and purified in DDM was identified by MALDI-TOF mass spectrometry (SI Appendix, Fig. S3). The addition of PEA to N. flavescens lipid A resulted in an increased cytokine response from THP-1 cells (SI Appendix, Fig. S4) (25–27). These results confirmed that the recombinant DDM solubilized enzyme was enzymatically active and able to induce a cytokine response.
Overview of Structural Fold.
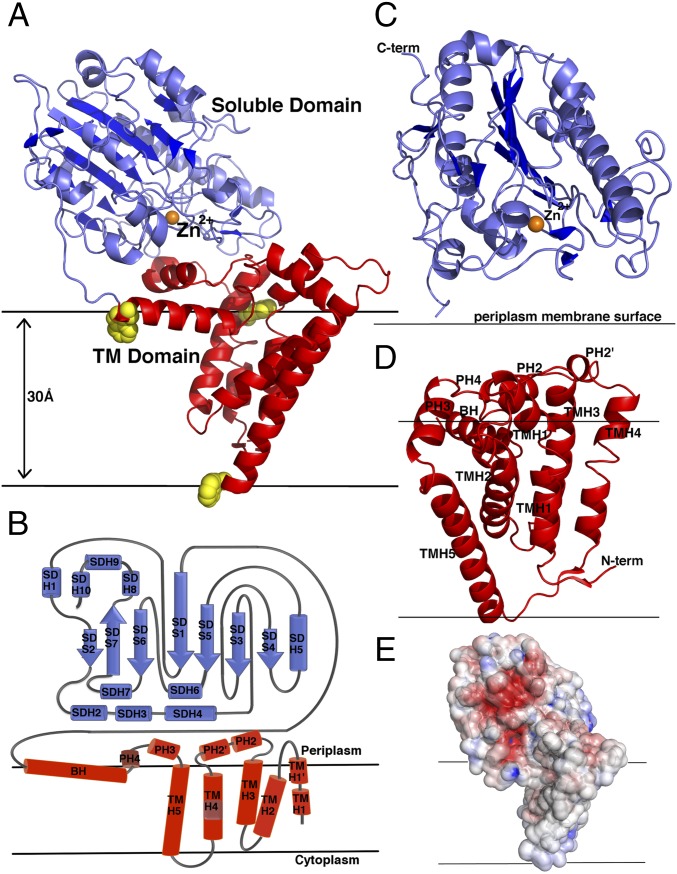
The crystal structure of the full-length NmEptA, was determined by molecular replacement, using the previously determined truncated periplasmic soluble domain (PDB code: 4KAV) as a search model for phasing (14). The structure is composed of two discretely folded domains: an N-terminal transmembrane (TM) domain and a C-terminal soluble periplasmic-facing domain (Fig. 1 A and B) connected by a bridging helix and an extended loop.
Fig. 1.
Molecular structure of NmEptA. (A) Ribbon representation of NmEptA. The amino-terminal TM domain is shown in red, and the carboxyl-terminal soluble domain is shown in blue. The side chains of the three TM domain tryptophan residues (Trp126, Trp148, and Trp207) are shown as yellow spheres. (B) The topology diagram of NmEptA showing the likely positioning of the protein relative to the periplasmic membrane with the coloring as shown in A. (C) Secondary structure of the soluble domain and (D) TM domain with the helical numbering labeled. (E) Electrostatic surface representation of NmEptA, calculated using APBS. The surface is color-contoured from −4 kT/e to +4 kT/e (negative in red, positive in blue). The bound Zn2+ ion in A and C is shown as an orange sphere. To delineate the proposed orientation of the protein within the bilayer, the membrane surface, representing the hydrophobic portion of the bilayer, is drawn as horizontal black lines.
The membrane domain contains five transmembrane helices (TMH1–TMH5; see Fig. 1 B and D) oriented approximately parallel to one another and spanning the inner membrane in a previously uncharacterized fold. The membrane domain contains two tryptophan residues and a number of tyrosine and histidine residues forming an aromatic belt along the membrane surface. These residues are likely to contribute to the stabilization and orientation of the protein in the bilayer and allow prediction of the membrane spanning regions of NmEptA (28). Intriguingly, of the five TM helices, only TMH5 spans the membrane with a length of 35 Å; the other four TM helices are shorter than the typical width of a membrane bilayer (30 Å).
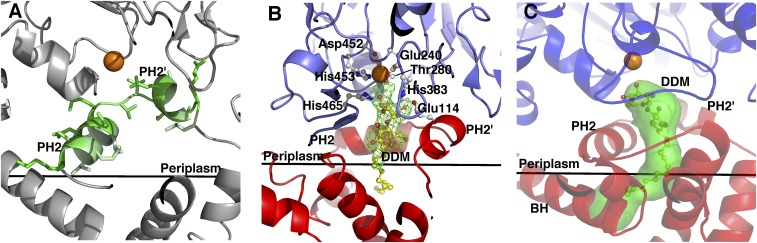
A patch of positively charged residues (Lys142, Lys144, Arg146, and Lys150) at the cytoplasmic end of TMH5 are likely to provide interactions with the phospholipid head groups at the inner membrane surface (Fig. 1E), as has been observed in other integral membrane proteins (29). TMH3 and TMH4 are positioned in close proximity to each other and are linked by a long loop at the periplasmic side, which harbors two small periplasmic facing helices, PH2 and PH2′ (Fig. 1 B and D). The hydrophobic representation of the NmEptA structure reveals PH2 and PH2′ helices with nonpolar residues aligned toward the membrane and polar residues toward the periplasm (Fig. 2A). The amphipathic nature of these helices suggests that they could be partially buried or embedded in the membrane, as has been reported in a tyrosine-interacting protein (30, 31). The central section of TMH4 adopts a π-helix configuration (Fig. 1D), and is linked to the TMH5 by a loop.
Fig. 2.
Active site region of NmEptA. (A) Amphipathic helices PH2 and PH2′, with the polar residues shown in dark green and nonpolar residues shown in pale green. (B) Difference electron density contoured at 3 σ, with a model of DDM included in the density. (C) Ribbon representation of NmEptA showing the tunnel (green) in the structure, as calculated by CAVER. The Zn2+ ion is represented as an orange sphere. In B and C, the TM domain is colored as a red ribbon representation, and the soluble domain is colored as a blue ribbon representation. The active site residues are shown as ball and stick representation colored with white bonds, and the DDM molecule is colored with yellow bonds. The membrane surface is shown by a black line in each panel.
TMH5 is the longest helix in the membrane domain, and based on the positions of the aromatic belt residues, its orientation may be tilted in the membrane. TMH5 is connected to the soluble domain by an extended periplasmic loop followed by a bridging helix. This extended periplasmic loop harbors two more periplasmic helices: the amphipathic PH3 and PH4, which adopts a 310 configuration.
The bridging helix (residues 194–208) has a highly unusual configuration in the structure. The N-terminal portion of the helix contains hydrophobic residues, whereas the C-terminal end is rich in charged residues. In addition, the C-terminal end contains two residues (Tyr204 and Trp207) that are positioned along the proposed membrane surface and comprise part of the aromatic belt. The nature of this helix suggests it may be tilted relative to the membrane surface and partially buried in the bilayer. An extended coiled region (residues 210–231) links the C terminus of the bridging helix with the start of the soluble domain.
The soluble domain adopts a hydrolase-type fold with a bound Zn2+ ion at the enzyme active site near to the catalytic nucleophile, Thr280 (Fig. 1C). The Zn2+ ion is bound identically to that seen in the truncated domain structure, and the intramolecular disulfide bonds are also preserved (14). A superposition of the truncated structure and the soluble domain in the full-length enzyme structures revealed no significant differences. Sequence comparisons with other PEA transferases acting on lipid A indicate that three of the five disulfide bonds (namely, Cys276-286, Cys327-Cys331, and Cys402-Cys410) are structurally conserved (SI Appendix, Fig. S1).
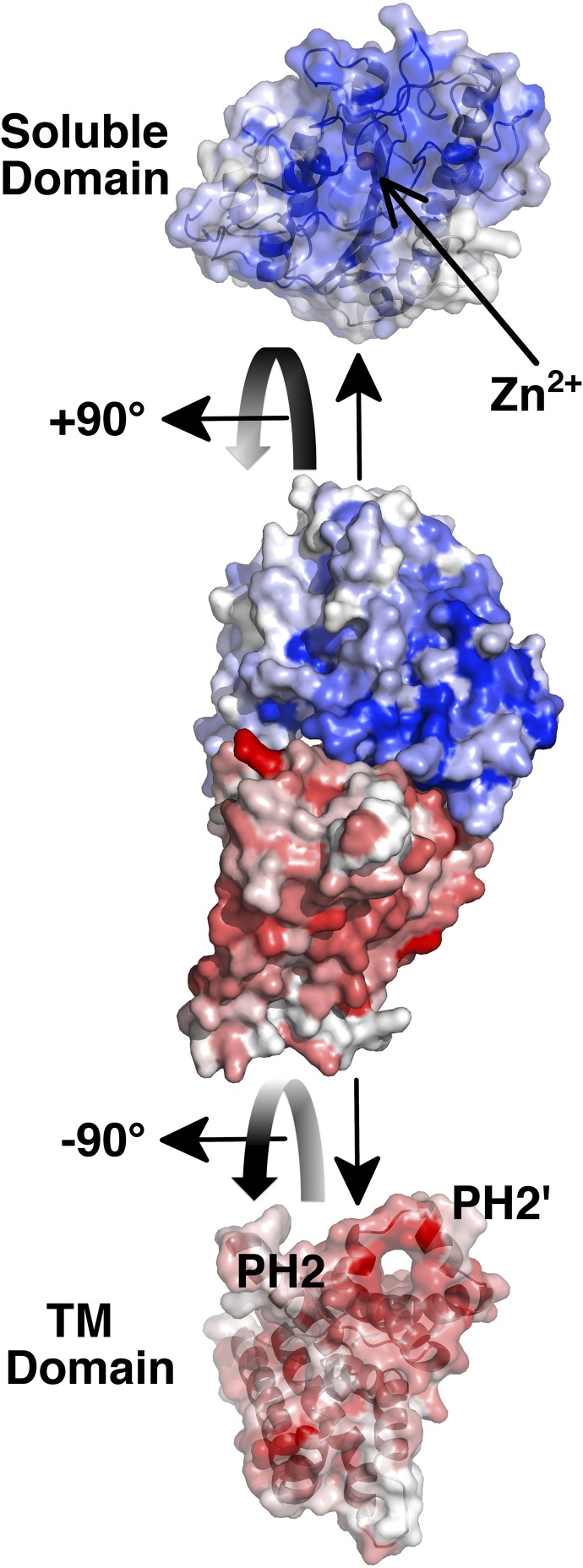
There is large interface between the soluble and membrane domains. According to an analysis with the server PDBePISA, this interdomain surface buries a region of ∼1,200 Å2. High conservation at the interface between the domains (Fig. 3) is revealed when the full-length structure is colored on the basis of sequence similarity with lipid A PEA transferases (SI Appendix, Fig. S1).
Fig. 3.
Surface area representation of NmEptA for the full-length structure and for the soluble domain and the TM domain. The colored shading of the surfaces corresponds to the different domains, as indicated. Each domain has been rotated from the view of the full-length structure to show the interaction surface. The intensities of the red and blue colors correspond to the level of sequence conservation, as indicated in the sequence alignment in SI Appendix, Fig. S1. The position of the PH2 and PH2′ helices in the TM domain and the position of the bound Zn2+ ion in the soluble domain are indicated.
The Active Site of NmEptA.
On completion of the modeling of the protein chain into the electron density map, residual difference electron density was present in the region of the active site and located between the PH2 and PH2′ helices, suggesting the presence of a bound ligand (Fig. 2A). A single molecule of DDM was modeled into this electron density and refined successfully. The first carbohydrate moiety of DDM lies deep in the substrate pocket, with the O3B hydroxyl group positioned 3.72 Å from the bound Zn2+ atom and 2.96 Å from the OH group of Thr280, the active site nucleophile. Three hydroxyl groups on the carbohydrate moiety of the DDM form hydrogen bonds with three invariant residues in the PH2 and PH2′ helices (Fig. 3 and SI Appendix, Fig. S1). The density for the more distal portion of the DDM acyl chain is weaker and the temperature factors become progressively larger compared with the carbohydrate moieties of the ligand, suggesting it does not bind strongly to any portion of the protein.
The truncated structure (14) revealed an exposed active site, and the surface comprising this region is likely to be orientated toward the membrane. This proposed orientation would allow direct access of the active site to the lipid substrates situated within membrane bilayer. The full-length structure confirms this prediction as the catalytic region of the enzyme faces toward the membrane domain and includes the PH2 and PH2′ helices.
This structural arrangement of the two domains in the presence of a bound ligand reveals a much more sequestered active site than previously observed for the truncated structure. In particular, the PH2 and PH2′ helices complete the substrate binding site, which extends toward the membrane domain, and presumably into the bilayer (Fig. 2B). Sequestering the active site is likely to be critical for efficient catalysis.
A further comparison of the two structures showed that O3B of DDM in the full-length protein is positioned identically to an oxygen atom of the phosphate moiety covalently bound to Thr280 in the truncated structure. Closer examination of the DDM binding mode reveals that the length of an enzyme-linked PEA molecule can be accommodated between Thr280 and Glu114, both of which are invariant between lipid A PEA transferases (SI Appendix, Fig. S1). Glu114, located in the PH2 helix, may hydrogen bond with the amine of PEA. Further analysis of this region of the structure was carried out using CAVER (32) and indicated a buried cavity (Fig. 2C). The size of this cavity precludes binding of a large lipid substrate such as lipopolysaccharide.
Conformational Flexibility.
As the crystal structure was determined using protein solubilized in DDM micelles, and a bound DDM molecule was visible near the active site, it was assumed that the bound detergent acts to hold the soluble domain close to the membrane domain, locking the enzyme in a particular conformation that does not accommodate a large lipid substrate.
To establish possible conformational flexibility of NmEptA, limited proteolysis and intrinsic tryptophan fluorescence experiments were undertaken. A study of the integral membrane enzyme PhoPQ-activated gene P (PagP) used an NMR-based approach in detergent environments to study the dynamics of the protein and revealed a more mobile state allowing substrate binding and a more rigid state enabling enzyme activity (33). Hence, detergent micelles can be used to sample conformational flexibility in membrane proteins.
NmEptA solubilized and purified in three different types of detergents, DDM, 6-cyclohexyl-1-hexyl-β-d-maltoside (Cymal-6), and Fos-choline-12 (FC-12), was used to study the conformational states of the protein. The protein integrity in the three detergents was assessed by circular dichroism (CD) spectroscopy (SI Appendix, Fig. S5A), which revealed that in all three detergents, NmEptA has secondary structure. In addition, the NBD-PEA TLC assay (SI Appendix, Fig. S2) was used to assess activity of the enzyme in different detergents. Finally, protein quality and thermal stability in DDM and FC-12, assessed by size exclusion chromatography-multiangle light scattering and differential scanning fluorometry (SI Appendix, Fig. S5 B and C), showed that the proteins are monodispersed and exhibit high thermal stability. The TLC assay showed that protein in detergents containing maltoside head groups (DDM and Cymal-6) were able to successfully remove PEA from the lipid substrate. In contrast, the enzyme in FC-12, a detergent containing a positively charged quaternary amine head group, failed to show product formation, suggesting the enzyme adopts a conformation that is not conducive to hydrolysis of PEA from a lipid substrate, although the overall secondary structure is maintained and the protein is stable.
Limited proteolysis profiles from trypsin and chymotrypsin digests of DDM, Cymal-6, and FC-12 purified enzyme (SI Appendix, Fig. S6) indicated differences in the rate of cleavage of the full-length protein between the three detergents. Proteins purified in DDM and Cymal-6 exhibited higher resistance to proteolytic cleavage, whereas protein purified in FC-12 showed a lower level of resistance to proteolysis.
Peptide analysis of the proteolytic fragments for the DDM and FC-12 purified proteins identified products containing the PH4 helix, the bridging helix, and the soluble domain (SI Appendix, Table S2). Analysis of the 42-kDa cleavage products by chymotrypsin revealed that the DDM-purified enzyme was cleaved at residue Tyr214 and the FC-12-purified enzyme cleaved at residue Tyr148, suggesting the enzyme adopts different conformational states in DDM and FC-12 micelles.
Further probing of possible conformational states involved intrinsic fluorescence studies that mapped the relative positions of the six tryptophan residues: Trp126 and Trp148 in the membrane domain, Trp207 in the bridging helix, and Trp247, Trp320 and Trp484 in the soluble domain. An analysis of buffer-corrected fluorescence emission profiles (SI Appendix, Fig. S7A) showed that the maximum (max) of the emission signal is red shifted for enzyme purified in FC-12 (emission peak max = 342 nm), relative to that in DDM or Cymal-6 (emission peak max = 336 nm). This red shift in FC-12 indicates a higher level of solvent exposure of tryptophan residues, which, considering the similar CD spectra for the three samples, suggests different conformations are adopted in different detergents. When NmEptA was denatured, the emission peaks had equivalent values, regardless of the detergent used for purification (SI Appendix, Fig. S7B). Quenching of the solvent exposed tryptophan residues by iodine revealed a Stern-Volmer constant in both DDM and Cymal-6-purified NmEptA that is 56–60% lower than that of NmEptA in FC-12 (SI Appendix, Fig. S7C). This difference decreased when protein was denatured (SI Appendix, Fig. S7D). Thus, altered protein conformations in different detergents contribute to quenching. The presence of a maltoside containing detergent may, through its ability to bind to the active site, hold the enzyme in a specific conformation that decreases the overall solvent exposure of tryptophan residues. In contrast, the presence of a detergent with a charged headgroup may allow the enzyme to adopt a different conformational state in which the tryptophan residues are more solvent exposed.
NmEptA in the Lipid Bilayer.
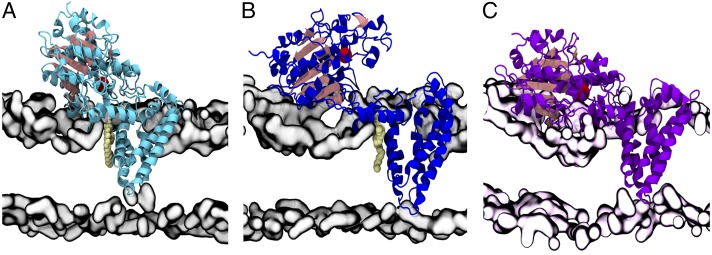
The relative orientation of NmEptA in the lipid bilayer, and the structural stability of the membrane-embedded protein, was investigated using molecular dynamics simulations. Because of the high percentage of PE lipids in Neisserial membranes (34), and the role of PE lipids as the first substrate in the reaction catalyzed by NmEptA, dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) lipids were used as a simplified representation of the Neisserial membrane in the initial simulation studies at both 298 K and 310 K. Additional simulations (298 K) were conducted in a PE bilayer enriched with 20% PG lipids to better reflect the physiological composition of the Neisserial membrane. Simulations of DPPE-embedded NmEptA showed the overall conformation of NmEptA remained close to the starting structure in four of the six simulations, with an average backbone rmsd of 3.3 ± 0.6 Å (SI Appendix, Fig. S8A) attributed to small changes in the relative orientation of the domains (Fig. 4A and SI Appendix, Fig. S8C). However, in two simulations, the soluble domain dissociated from the membrane domain and “rolled” over the membrane surface, interacting with the lipid polar heads (Fig. 4B and SI Appendix, Fig. S8D). This movement gives an increase in the distance between the centers of mass of the domains with associated increases in the backbone rmsd (SI Appendix, Figs. S9A and 10A), and induces changes in the local curvature of the bilayer in the interaction area (Movies S1 and S2). In the PE/PG membrane, the conformation of NmEptA was similar to those obtained in the DPPE membrane. Here, the distance between the center of mass of the TM and the soluble domains was 5–10 Å greater than observed in the closed DPPE-embedded NmEptA conformations (SI Appendix, Fig. S10), suggesting the protein can adopt a range of conformations. The molecular interactions triggering the conformational change remain unclear; however, positively charged residues dispersed on the surface of the soluble domain play an important role in the steering and binding of the soluble domain to the membrane surface (Movies S1 and S2).
Fig. 4.
Molecular dynamics simulations of NmEptA in a DPPE and PE/PG membrane environment. (A) Representative closed conformation of NmEptA embedded in DPPE bilayer obtained after 100-ns simulation at 310 K with a bound DPPE molecule positioned between periplasmic helices PH2 and PH2′. (B) Representative open conformation of NmEptA embedded in DPPE bilayer after 100-ns simulation at 310 K with the bound DPPE lipid. (C) Representative conformation of NmEptA embedded in PE/PG bilayer after 100-ns simulation at 298 K. The bound DPPE molecules are shown in pale yellow spheres, and the Zn2+ ion is shown as an oversized red sphere. The polar region of the bilayer is represented by a gray surface defined by phosphorus and nitrogen atoms for the DPPE bilayer and by a pale pink surface for the mixed PE/PG bilayer.
In one simulation of the DPPE-embedded NmEptA a single PE headgroup spontaneously bound between the PH2 and PH2′ helices (Fig. 4 A and B) and remained stably bound throughout the remainder of the simulation. The high sequence conservation of PH2 and PH2′ (SI Appendix, Fig. S1), together with their position at the membrane surface, suggests a key role in substrate binding.
Comparison Between NmEptA and ArnT.
Recently, the structure of the lipid-to-lipid glycosyltransferase 4-amino-4-deoxy-l-arabinose transferase (ArnT) has been determined (35). ArnT is an integral membrane bound glycosyltransferase that attaches the carbohydrate, 4-amino-4-deoxy-l-arabinose (l-Ara4N) to the 1 and 4′ phosphate groups of lipid A, the same site of modification for EptA. In the case of ArnT, however the donor substrate is undecaprenyl phosphate-α-l-Ara4N. Both enzymes are located in the periplasmic membrane and contain a TM domain and a periplasmic-facing soluble domain. Although the enzymes modify the same site on lipid A, their structures are distinctly different from one another. The N-terminal TM domain of ArnT is considerably larger, comprising 13 TM helices, whereas the C-terminal soluble periplasmic-facing portion of the enzyme adopts a smaller domain structure comprising ∼90 residues. The observed differences in the structures of the two enzymes are likely to be a result of the different donor substrates required (PE in the case of NmEptA and sugar-based lipid in the case of ArnT). These structural differences may also reflect different mechanisms of catalysis for each enzyme. In ArnT, both the substrates are required to bind to the enzyme to enable direct transfer of l-Ara4N from undecaprenyl phosphate to lipid A. The structure of the enzyme reveals three cavities proposed to house the lipid substrates lipid A and undecaprenyl phosphate-α-l-Ara4N. Two of the cavities are occupied by undecaprenyl phosphate in a complex structure with the ligand. The third cavity is proposed to house the lipid A substrate, and it is proposed that this cavity becomes accessible through the binding of the donor substrate. In contrast, NmEptA involves the formation of a Thr280-PEA enzyme intermediate similar to that observed for other alkaline phosphatase-type phosphate transferases, hence suggesting a ping-pong mechanism.
In summary, the crystal structure of EptA from Neisseria reveals a protein fold with a membrane-bound domain and a periplasmic-facing soluble domain. The substrate binding pocket involves the soluble domain, as well as the PH2 and PH2′ helices on the membrane domain. Sequence conservation among lipid A PEA transferases further highlights the importance of this region in substrate recognition and recruitment.
Experimental techniques including intrinsic fluorescence and limited proteolysis suggest the protein is able to adopt different conformational states. Molecular dynamics provides intriguing insights into a potential conformational change that the enzyme may undergo. On the basis of these observations, we propose that a conformational change may be necessary for the binding of two differently sized lipid substrates to facilitate transfer of PEA, possibly by a ping-pong mechanism.
Antivirulence therapy, in which virulence mechanisms of a pathogen are chemically inactivated, represents a promising approach to the development of treatment options. EptA is one such virulence-inducing target in Neisseria and is homologous to the recently identified colistin resistance enzyme, Mcr-1, found on a transferable plasmid. As this enzyme is a validated target for treatment of MDR gram-negative bacterial infections, this structure provides a strong basis for a structure-guided approach to develop small molecule inhibitors to combat multidrug resistance in pathogenic gram-negative bacteria. Clearly, further studies are required to characterize the different states of the enzyme, which might reveal alternative binding sites for inhibitors. Critically, the structure presented here extends our understanding of the substrate-binding site of EptA by including contributions from both membrane and soluble domains.
Materials and Methods
NmEptA was expressed as a hexahistidine-tagged protein in E. coli cells. The enzyme was purified in the presence of DDM by Ni2+ NTA affinity and size exclusion chromatography. Crystals were obtained and the structure solved by molecular replacement, using the soluble domain structure (PDB accession code: 4KAV) (14). The data collection, processing, and refinement statistics are given in the SI Appendix, Table S1. The reaction of N. flavescens lipid A of lipooligosaccharide with NmEptA was followed by MALDI-TOF mass spectrometry. In addition, an enzyme assay was carried out on NmEptA purified in different detergent micelles (DDM, FC-12, and Cymal-6), using a substrate containing a fluorescent label, NBD-PEA. Finally a TNFα assay was carried out using human monocytic leukemia cell THP-1 cells. For limited proteolysis experiments, NmEptA purified in different detergents (DDM, FC-12, and Cymal-6) were treated with trypsin or chymotrypsin over the course of 24 h, and the resultant cleaved fragments were analyzed by MALDI-TOF mass spectrometry. Tryptophan intrinsic fluorescence of NmEptA in different detergents and under denaturing conditions was measured. Quenching experiments were undertaken using potassium iodide. Molecular dynamics simulations were carried out using GROMACS (36) version 3.3.3 with the GROMOS 54A7 force field (37). Full methods and accompanying references are available in the SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dongxiao Feng for his technical assistance, Prof. Francis C. Dzoka (Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco) for access to the Microflex LT mass spectrometer, and the beamline scientists at the Diamond Light Source and the Australian Synchrotron for support during X-ray data collection. This work was supported by National Health and Medical Research Council of Australia Grants APP1003697 (to A.V. and C.M.K.), APP1078642 (to A.V., C.M.K., and K.A.S.), and APP1049685 (to M.L.O.); the Research Service of the US Department of Veterans Affairs Merit Review Award BX000727 (to G.A.J.); and the Welcome Trust Grant 099165/Z/12/Z (to Prof. S. Iwata and I.M.). We acknowledge the Australian Research Council for Infrastructure Grant LE120100092 (to A.V. and K.A.S.) and Fellowship Awards FT100100291 (to K.A.S.) and DE120101550 (to M.L.O.). A.A. and S.S.W. acknowledge the University of Western Australia for support through Australian Postgraduate Awards, a Fay Gale Fellowship, and a Bruce and Betty Green Scholarship. This work used resources from the National Computational Infrastructure, supported by the Australian Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.v.d.B. is a Guest Editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5FGN). The molecular dynamics simulations have been deposited to FigShare, www.figshare.com, and are available as DOI: 10.6084/m9.figshare.4595473.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612927114/-/DCSupplemental.
References
- 1.World Health Organization . Antimicrobial resistance: Global report on surveillance. WHO; Geneva: 2014. pp. 1–256. [Google Scholar]
- 2.US Department of Health and Human Services . Antibiotic resistance threats in the United States 2013. Centers for Disease Control and Prevention; Atlanta: 2013. pp. 1–113. [Google Scholar]
- 3.Osei Sekyere J, Govinden U, Bester LA, Essack SY. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: Emerging resistance mechanisms and detection methods. J Appl Microbiol. 2016;121(3):601–617. doi: 10.1111/jam.13169. [DOI] [PubMed] [Google Scholar]
- 4.Liu YY, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 5.McGann P, et al. Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF Plasmid: First Report of mcr-1 in the United States. Antimicrob Agents Chemother. 2016;60(7):4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rapoport M, et al. First description of mcr-1-mediated colistin resistance in human infections caused by Escherichia coli in Latin America. Antimicrob Agents Chemother. 2016;60(7):4412–3. doi: 10.1128/AAC.00573-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pristovsek P, Kidric J. Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: An NMR and molecular modeling study. J Med Chem. 1999;42(22):4604–4613. doi: 10.1021/jm991031b. [DOI] [PubMed] [Google Scholar]
- 8.Needham BD, Trent MS. Fortifying the barrier: The impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol. 2013;11(7):467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartley SN, Kahler CM. The glycome of Neisseria spp.: How does this relate to pathogenesis? In: Kahler CM, Davies JK, editors. Pathogenic Neisseria: Genomics, molecular biology, and disease intervention. Caister Academic Press; Poole, UK: 2014. pp. 114–145. [Google Scholar]
- 10.Cox AD, et al. Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: Identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide. J Bacteriol. 2003;185(11):3270–3277. doi: 10.1128/JB.185.11.3270-3277.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperandeo P, et al. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol. 2007;189(1):244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzeng YL, et al. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J Bacteriol. 2005;187(15):5387–5396. doi: 10.1128/JB.187.15.5387-5396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobbs MM, et al. Lipid A’s structure mediates Neisseria gonorrhoeae fitness during experimental infection of mice and men. MBio. 2013;4(6):e00892–13. doi: 10.1128/mBio.00892-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wanty C, et al. The structure of the neisserial lipooligosaccharide phosphoethanolamine transferase A (LptA) required for resistance to polymyxin. J Mol Biol. 2013;425(18):3389–3402. doi: 10.1016/j.jmb.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Fage CD, Brown DB, Boll JM, Keatinge-Clay AT, Trent MS. Crystallographic study of the phosphoethanolamine transferase EptC required for polymyxin resistance and motility in Campylobacter jejuni. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 10):2730–2739. doi: 10.1107/S1399004714017623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stojanoski V, et al. Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC Biol. 2016;14(1):81. doi: 10.1186/s12915-016-0303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cullen TW, Madsen JA, Ivanov PL, Brodbelt JS, Trent MS. Characterization of unique modification of flagellar rod protein FlgG by Campylobacter jejuni lipid A phosphoethanolamine transferase, linking bacterial locomotion and antimicrobial peptide resistance. J Biol Chem. 2012;287(5):3326–3336. doi: 10.1074/jbc.M111.321737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cullen TW, et al. EptC of Campylobacter jejuni mediates phenotypes involved in host interactions and virulence. Infect Immun. 2013;81(2):430–440. doi: 10.1128/IAI.01046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullen TW, Trent MS. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc Natl Acad Sci USA. 2010;107(11):5160–5165. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott NE, et al. Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J Biol Chem. 2012;287(35):29384–29396. doi: 10.1074/jbc.M112.380212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Loo B, et al. An efficient, multiply promiscuous hydrolase in the alkaline phosphatase superfamily. Proc Natl Acad Sci USA. 2010;107(7):2740–2745. doi: 10.1073/pnas.0903951107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boltes I, et al. 1.3 A structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family. Structure. 2001;9(6):483–491. doi: 10.1016/s0969-2126(01)00609-8. [DOI] [PubMed] [Google Scholar]
- 23.Lukatela G, et al. Crystal structure of human arylsulfatase A: The aldehyde function and the metal ion at the active site suggest a novel mechanism for sulfate ester hydrolysis. Biochemistry. 1998;37(11):3654–3664. doi: 10.1021/bi9714924. [DOI] [PubMed] [Google Scholar]
- 24.von Bülow R, Schmidt B, Dierks T, von Figura K, Usón I. Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis. J Mol Biol. 2001;305(2):269–277. doi: 10.1006/jmbi.2000.4297. [DOI] [PubMed] [Google Scholar]
- 25.John CM, Liu M, Jarvis GA. Profiles of structural heterogeneity in native lipooligosaccharides of Neisseria and cytokine induction. J Lipid Res. 2009;50(3):424–438. doi: 10.1194/jlr.M800184-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.John CM, Liu M, Jarvis GA. Natural phosphoryl and acyl variants of lipid A from Neisseria meningitidis strain 89I differentially induce tumor necrosis factor-alpha in human monocytes. J Biol Chem. 2009;284(32):21515–21525. doi: 10.1074/jbc.M109.004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu M, John CM, Jarvis GA. Phosphoryl moieties of lipid A from Neisseria meningitidis and N. gonorrhoeae lipooligosaccharides play an important role in activation of both MyD88- and TRIF-dependent TLR4-MD-2 signaling pathways. J Immunol. 2010;185(11):6974–6984. doi: 10.4049/jimmunol.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez KM, Kang G, Wu B, Kim JE. Tryptophan-lipid interactions in membrane protein folding probed by ultraviolet resonance Raman and fluorescence spectroscopy. Biophys J. 2011;100(9):2121–2130. doi: 10.1016/j.bpj.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulmschneider MB, Sansom MS. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta. 2001;1512(1):1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 30.Segrest JP, De Loof H, Dohlman JG, Brouillette CG, Anantharamaiah GM. Amphipathic helix motif: Classes and properties. Proteins. 1990;8(2):103–117. doi: 10.1002/prot.340080202. [DOI] [PubMed] [Google Scholar]
- 31.Min CK, et al. Role of amphipathic helix of a herpesviral protein in membrane deformation and T cell receptor downregulation. PLoS Pathog. 2008;4(11):e1000209. doi: 10.1371/journal.ppat.1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chovancova E, et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput Biol. 2012;8(10):e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang PM, Bishop RE, Kay LE. The integral membrane enzyme PagP alternates between two dynamically distinct states. Proc Natl Acad Sci USA. 2004;101(26):9618–9623. doi: 10.1073/pnas.0402324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahman MM, et al. The membrane phospholipids of Neisseria meningitidis and Neisseria gonorrhoeae as characterized by fast atom bombardment mass spectrometry. Microbiology. 2000;146(Pt 8):1901–1911. doi: 10.1099/00221287-146-8-1901. [DOI] [PubMed] [Google Scholar]
- 35.Petrou VI, et al. Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science. 2016;351(6273):608–612. doi: 10.1126/science.aad1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 37.Schmid N, et al. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J. 2011;40(7):843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.