Fig. 4.
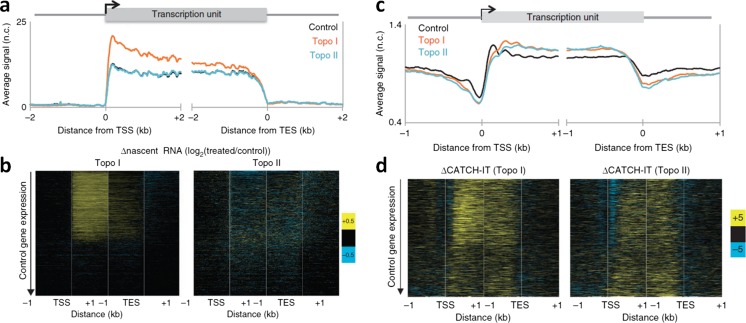
Transcription-generated torsional stress can destabilize nucleosomes. a, b Altered Pol II elongation kinetics upon Topo I or Topo II inhibition. a The average transcription profiles surrounding the transcription start sites (TSSs) and transcription end site (TESs) of all genes. Topo I inhibition increases the overall nascent-RNA production near TSSs, but not near TESs, suggesting Pol II stalling before the completion of the transcription, whereas Topo II inhibition showed no overall change relative to the control. b Heat maps of the log ratio of nascent RNA for Topo I inhibited samples over the control (left) and Topo II inhibited samples over the control (right), with genes ordered by decreasing expression in control samples. Topo I inhibition primarily affects transcribed genes, whereas Topo II inhibition results in heterogeneous changes in nascent-RNA levels, suggesting that Topo II plays only a secondary role in transcription. c, d Altered nucleosome turnover under torsion. c Nucleosome turnover is measured by CATCH-IT, followed by mapping of the positions of captured nucleosomes. d Heat maps showing changes in CATCH-IT signals, after Topo I (left) and Topo II (right) inhibition, from the controls. Adapted from Teves and Henikoff (2014) with permission