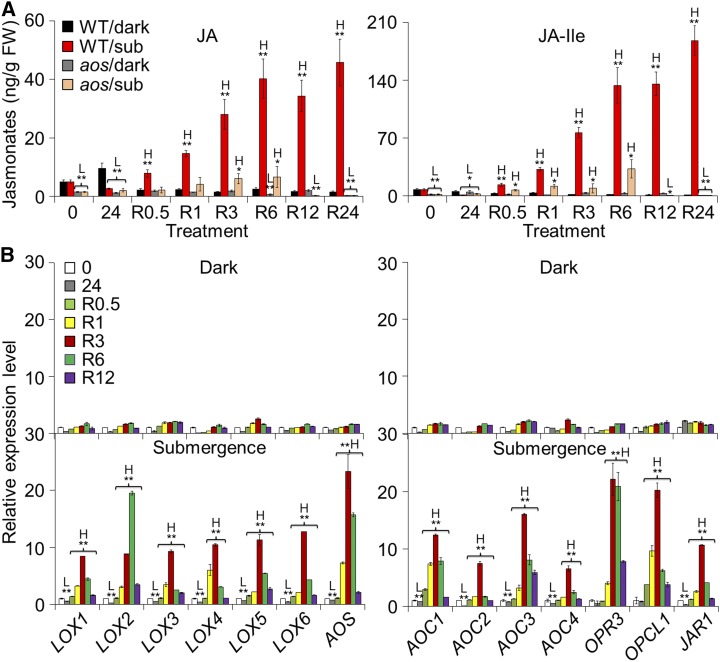
Figure 1.
Induction of JA biosynthesis by reoxygenation. A, The endogenous JA levels in the wild type (WT) and aos mutant in response to submersion and reoxygenation. Four-week-old WT and aos mutant plants were treated with dark only or dark submergence for 24 h followed by reoxygenation. Rosette leaf samples were collected for JA extraction at 0 and 24 h of dark or dark submergence as well as 0.5, 1, 3, 6, 12, and 24 h of reoxygenation and then analyzed by liquid chromatography/mass spectrometry. H2-JA was added as an internal quantitative standard. Two biological replicates were conducted with similar results, and the representative data from one replicate is shown. The data are means ± sd (n = 8 technical replicates; 200 mg of leaves harvested from three independent plants were pooled for each technical replicate). B, Quantitative PCR analyses showing the reoxygenation-inducible expression of JA biosynthetic genes (LOX1, LOX2, LOX3, LOX4, LOX5, LOX6, AOS, AOC1, AOC2, AOC3, AOC4, OPR3, OPCL1, and JAR1). Total RNA was isolated from rosettes of 4-week-old soil-grown plants at 0 and 24 h of dark or dark submergence at 0.5, 1, 3, 6, and 12 h of reoxygenation. Transcript levels relative to 0 h (prior to treatment) for each time point were normalized to the levels of TUB3. Three biological replicates were conducted with similar results, and the representative data from one replicate is shown. The data are means ± sd (n = 3 technical replicates). Asterisks with “H” or “L” indicate significant higher or lower than that of WT (*P < 0.05; **P < 0.01 by Student’s t test).