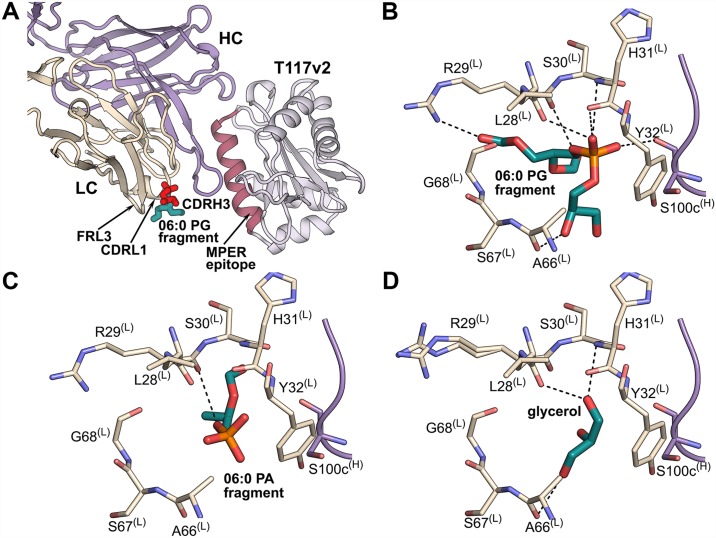
Fig 1. 10E8 lipid-binding site.
(A) Crystal structure of the 10E8-T117v2 complex bound to 06:0 PG (sticks; red, glycerol and phosphate moieties of the head group; cyan, head group region linked to the lipid tails). The 10E8 light chain (LC) is shown in beige, heavy chain (HC) in violet, and T117v2 scaffold containing the MPER epitope (pink) in gray. This color scheme is used throughout. (B) All potential hydrogen-bond interactions (within ~3.5 Å) of the 06:0 PG fragment (colored by atoms) with 10E8 residues of CDRL1 (beige) and CDRH3 (violet) are shown as dashed lines. (C) All potential hydrogen-bond interactions (within ~3.5 Å) of the 06:0 PA fragment (colored by atoms) with CDRL1 (beige) and CDRH3 (violet) residues of 10E8. (D) All potential hydrogen-bond interactions (within ~3.5 Å) of a glycerol found in the lipid-binding site in the 10E8-T117v2 structure in the absence of added lipids.