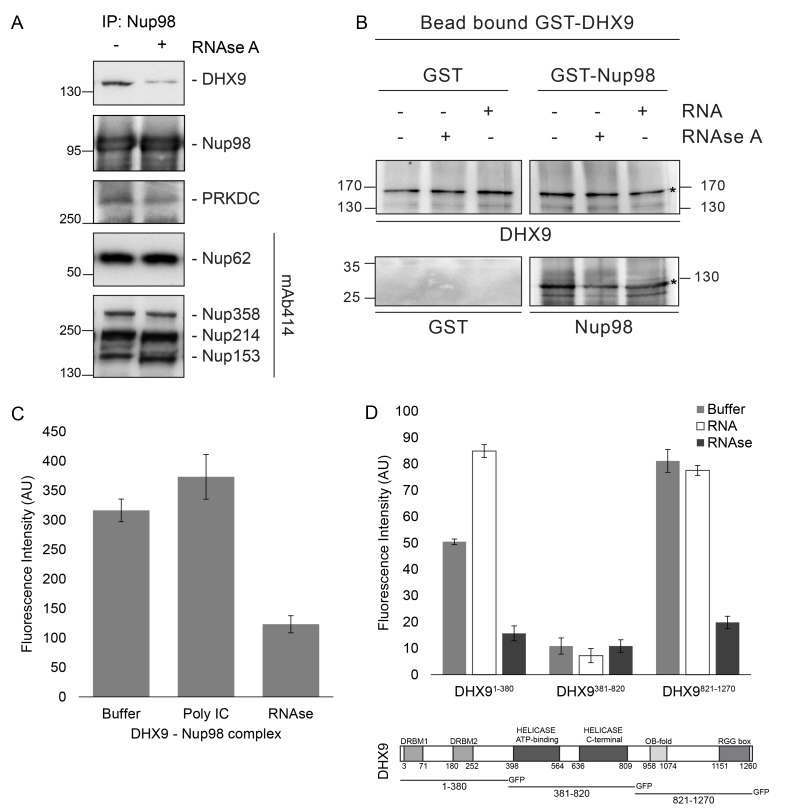
Figure 6. Nup98 binds directly to DHX9.
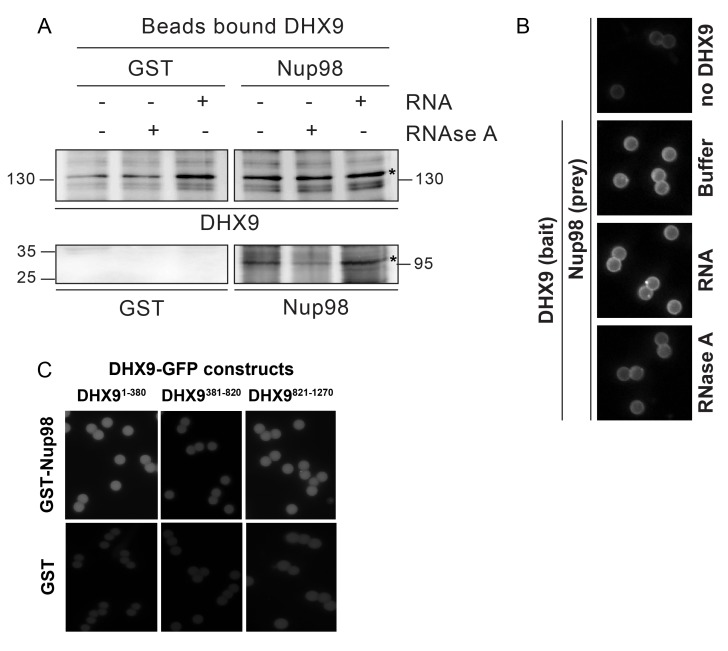
(A) Nup98 was affinity purified from HEK293T cell lysates. Bead-bound protein complexes were then incubated with or without RNase A, and proteins remaining bound to Nup98 were analyzed by western blotting using antibodies directed against the indicated proteins (right). The positions of molecular mass markers (shown in kDa) are indicated on the left. (B) Anti-DHX9 antibodies coupled to beads were used to immobilize GST-DHX9. Bead-bound GST-DHX9 was incubated with GST-Nup98 or GST alone in the presence of RNA (poly I:C), RNase A, or buffer alone. Bound proteins were analyzed by western blotting using the indicated antibodies (below each panel). The top row of images shows the GST-DHX9 bait bound to beads. The bottom row of images shows GST and GST-Nup98 that bound to GST-DHX9 under the indicated conditions. Asterisks denote positions of GST-DHX9 and GST-Nup98. The positions of molecular mass markers (shown in kDa) are indicated. A similar interaction between untagged recombinant DHX9 and Nup98 was also detected (Figure 6—figure supplement 1A). (C) Bead halo assays were performed using DHX9 immobilized on beads with an anti-DHX9 antibody as bait and Nup98 as prey. Prior to the binding step, both proteins were incubated with RNase A, RNA (poly I:C), or buffer alone. Interactions of Nup98 with bead-bound DHX9 were detected by fluorescence microscopy with rabbit anti-Nup98 antibodies and Alexa Fluor 488 donkey anti-rabbit antibodies. Examples of images of beads used to quantify binding can be seen in Figure 6—figure supplement 1B. (D) Bead halo assays were performed using bead-bound GST-Nup98 (bait) and different domains of DHX9-GFP (prey; see bottom diagram). Proteins were incubated with RNA (poly I:C), RNase A, or buffer alone before binding. The interaction of bead-bound GST-Nup98 with DHX9-GFP domains was detected by fluorescence microscopy. Examples of images of beads can be seen in Figure 6—figure supplement 1C. For C and D, bead halo assays were performed with purified recombinant proteins. Plots show average fluorescence intensity values of beads (arbitrary units) corrected against negative control assays (see Materials and methods). Results from three biological replicates are shown. Error bars indicate standard deviation between biological replicates.