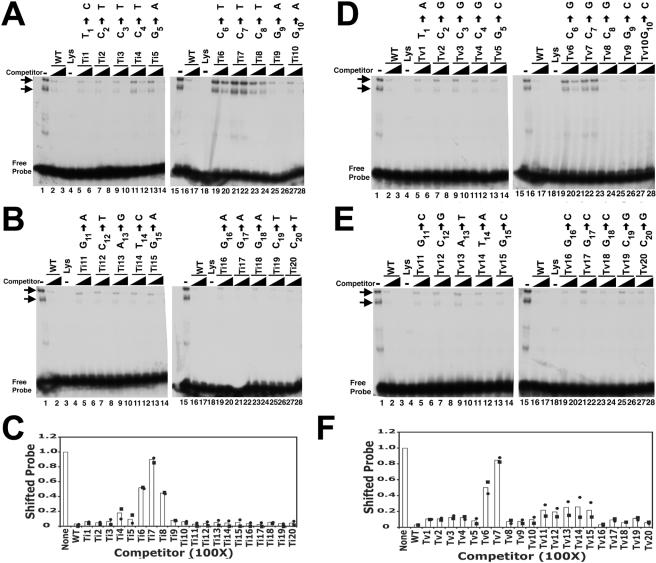
FIG. 2.
Competition of LANA1 complexes with oligonucleotides containing point mutations. (A to C) EMSA results after competition with oligonucleotides containing point transition mutations. (A and B) An EMSA was performed after the TR-13 radiolabeled probe was incubated with in vitro-translated LANA1 (lanes 1 and 15) or reticulocyte lysate (lanes 4 and 18). Unlabeled TR-13 (WT) or the indicated transition mutant (Ti1 through Ti20) at a 25- or 100-fold excess was included in the incubation mixture prior to the EMSA as indicated. The transition substitution in each mutant is indicated above the lanes. Horizontal arrows identify LANA1 complexes. Data shown are representative of three experiments. (C) Quantitation of EMSA results. The signals from two experiments (shown as circles and squares) were captured and averaged (bars). Data are from competition with oligonucleotides at a 100-fold excess. (D to F) EMSA results after competition with oligonucleotides containing point transversion mutations. (D and E) EMSA with the TR-13 radiolabeled probe after incubation with in vitro-translated LANA1 (lanes 1 and 15) or reticulocyte lysate (lanes 4 and 18). Unlabeled TR-13 (WT) or the indicated transversion mutant (Tv1 through Tv20) at a 25- or 100-fold excess was included in the incubation mixture prior to the EMSA. The transversion substitution for each mutant is indicated above the lanes. Arrows indicate LANA1 complexes. The data shown are representative of three experiments. (F) Quantitation of EMSA results. The signals from two experiments (shown as circles and squares) were captured and averaged (bars). Data are from competition with oligonucleotides at a 100-fold excess. (C and F) Values were determined by dividing the shifted signal after incubation with competitor oligonucleotide by the shifted signal in the absence of competitor.