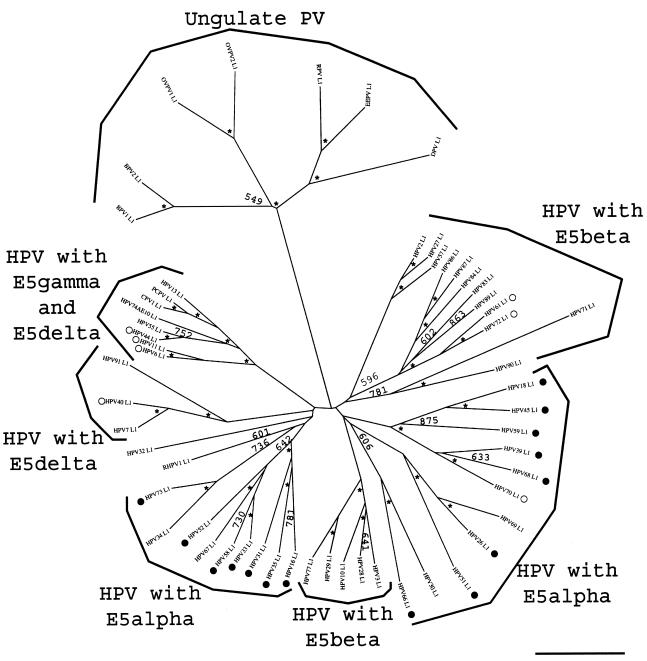
FIG. 5.
Phylogenetic tree of the L1 sequences in mucosal HPVs. Protein sequences were retrieved from the Los Alamos HPV sequence database or TrEMBL and aligned with the TCOFFEE algorithm. A phylogenetic tree was constructed from the multiple alignments by the maximum-likelihood method. Similar topologies were obtained when using DIALIGN and CLUSTAL W as alignment algorithms and neighbor-joining methods as phylogenetic methods. Numbers at the nodes indicate bootstrap support values after 1,000 bootstrap cycles (only values above 500 are given). An asterisk indicates that the bootstrap support is above 950. The bar at the bottom gives the relationship between branch lengths and 0.1 matrix units. High-risk and low-risk mucosal HPVs are labeled with black and white circles, respectively. L1 sequences in ungulate PVs are included as outgroups. An early split event separated mucosal HPV L1 proteins in two main branches, close to the divergence point with the ancestor from ungulate PV L1 proteins. The phylogenetic relationships, according to L1 proteins, do not match the epidemiological classification of these viruses. The phylogenetic relationships among viruses were the same as those for L1 and L2 but differed for E5, E6, and E7 proteins.