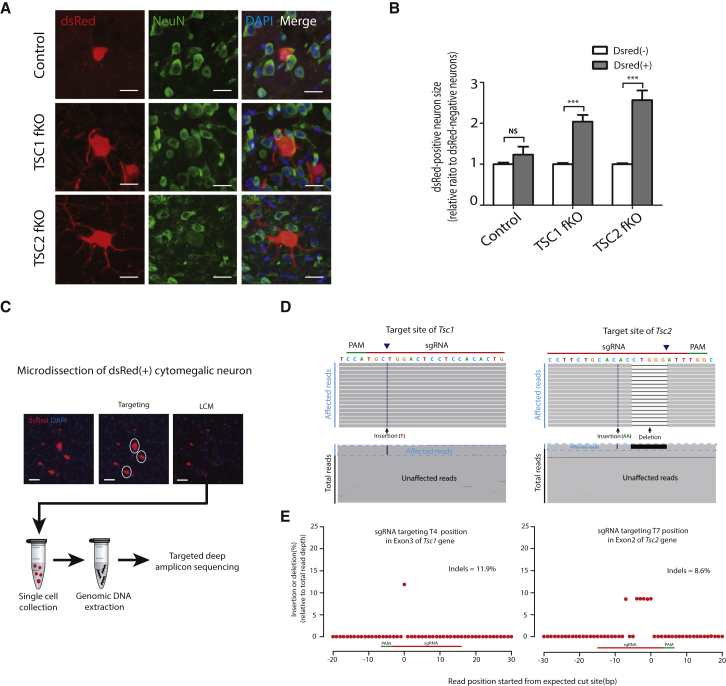
Figure 5.
Cytomegalic Neurons Contain the Correctly Edited Genome Sequence of Tsc1 and Tsc2 Targeted by the CRISPR-Cas9 Construct
(A) Co-immunostaining for NeuN and DAPI. NeuN staining of dsRed-positive cells is presented. The soma size of dsRed-positive neurons was measured. Scale bars, 20 μm.
(B) Bar chart of the neuronal size in each group shows an increased soma size in affected cortical regions. Data represent the mean ± SEM (n = 27–299 per group). ∗∗∗p < 0.001 compared with dsRed-negative neurons (Student’s t test).
(C) Schematic figure of the microdissection procedure used to isolate dsRed-positive cytomegalic neurons and subsequent targeted deep amplicon sequencing. The dsRed-positive cytomegalic neurons are denoted by a white circle in the LCM image (“targeting”). Labeled neurons were microdissected, and 10∼20 cells were collected in an adhesive cap tube. Genomic DNA was then extracted from the collected cells and submitted for deep amplicon sequencing. Scale bars, 50 μm.
(D) Representative view of Tsc1 and Tsc2 indels induced by Tsc1 and Tsc2 target sgRNA from the Integrative Genomic Viewer. Black and purple bars indicate deletions and insertions, respectively. The arrowheads indicate the theoretical cutting site of Cas9. PAM, protospacer adjacent motif.
(E) The frequency (relative to total reads) and position of indels are shown. The 0 position indicates the theoretical cutting site of Cas9 guided by sgRNA. The detailed amplicon sequencing procedure and deep sequencing data for the control group are presented in Figure S14.