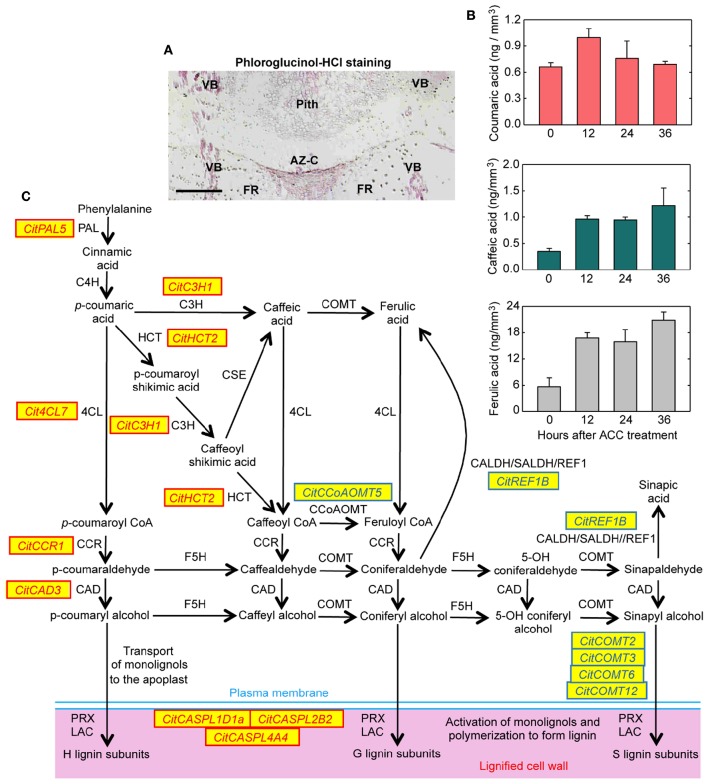
Figure 8.
Lignin biosynthesis and deposition in the abscission zone area during citrus fruit abscission. (A) Tissue localization of lignin through phloroglucinol-HCl staining in longitudinal sections of the AZ-C from Washington Navel fruits treated for 48 h with ethylene. Lignin is deposited at the central core of the AZ-C, at the separation line, and spreads to the adjacent cells of the fruit rind through the distal side of the AZ-C. Scale bar: 500 μm. Key labeling: AZ-C, abscission zone C; FR, fruit rind; VB, vascular bundles. (B) Lignin biosynthesis intermediates were quantified through UPLC-MS/MS in AZ-C cells at 0, 12, 24, and 36 h after ACC treatment. Data are expressed as ng of coumaric acid, caffeic acid and ferulic acid per mm3 of microdissected tissue. The results are means of three independent samples containing ~40,000 pooled AZ-C cells ±SE. (C) Genes belonging to the general phenylpropanoid and monolignol biosynthesis pathways and lignin polymerization up- or down-regulated exclusively in the fruit AZ-C cells during ethylene-promoted citrus fruit abscission. Enzymes and proteins associated with monolignol biosynthesis and polymerization are: phenylalanine ammonia lyase (PAL), trans-cinnamate 4-hydroxylase (C4H), 4-coumarate:CoA ligase (4CL), hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyl transferase (HCT), coniferaldehyde dehydrogenase/sinapaldehyde dehydrogenase (CALDH/SALDH), caffeoyl shikimate esterase (CSE), p-coumarate 3-hydroxylase (C3H), caffeoyl-CoA 3-O-methyltransferase (CCoAOMT), cinnamoyl-CoA reductase (CCR), ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), cinnamyl alcohol dehydrogenase (CAD), Casparian strip membrane domain protein-like (CASPL), laccase (LAC) and peroxidase (PRX).