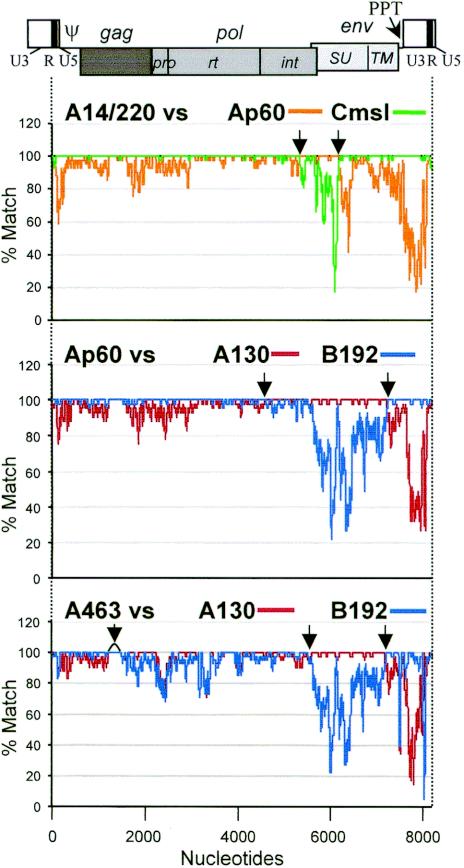
FIG. 1.
PERV recombination profiles shown by local homology analysis. (Top) Four full-length nucleotide sequences, A14/220 (8,188 nt), Cmsl (8,144 nt), Ap60 (8,261 nt), and A130 (8,299 nt), were analyzed by LOHA with 41-nt windows. The alignment length was 8,325 nt positions and a percent match plot for A14/220 versus Cmsl was overlaid on that for A14/220 versus Ap60. (Middle) Ap60, A130, and B192 (8,255 nt) were analyzed, and a percent match plot for Ap60 versus B192 was overlaid on that for Ap60 versus A130. The alignment size was 8,372 positions. (Bottom) A463 (8,204 nt), A130, and B192 were aligned: A463 versus B192 overlaid on A463 versus A130 for 8453 positions. A schema of a PERV provirus is displayed on top to indicate the positions of the viral genes and domains. The apparent recombination crossover points are indicated by arrows.