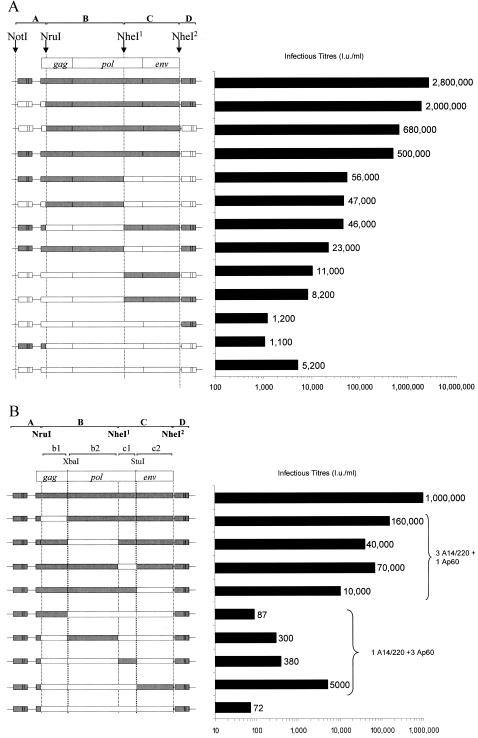
FIG. 5.
Infectious titers of A14/220 and Ap60 chimeras. (A) Full-length proviral PERV chimeras consisting of A14/220 (gray bars) or Ap60 (white bars) sequences were constructed by using four restriction sites: two conserved sites (NruI and NheI1 sites), an NheI2 site (created in Ap60 by mutagenesis), and an NotI site outside of the provirus genome. Infectious titer (infectious units per milliliter) of replicating PERV following establishment of persistently infected cultures by DNA transfection was measured on human 293 cells by an immunocytological focus assay (2). A data set of a representative of two independent experiments is shown by bars as well as numbers. (B) Chimeric PERV containing a mix-and-match set of protein coding region (fragments B and C in panel A) between A14/220 (gray bars) and Ap60 (white bars) sequences as well as the rest of the genome derived from A14/220 were constructed. Fragments B were divided to subfragments b1 and b2 by XbaI and fragments C were divided into to c1 and c2 by StuI. Results for chimeras containing three subfragments from A14/220 (three A14/220 plus one Ap60) as well as one subfragment from A14/220 (one A14/220 plus three Ap60) are shown. Infectious titer (infectious units per milliliter) of replicating PERV harvested 3 days after DNA transfection was measured on human 293 cells by an immunocytological focus assay. A data set of a representative of two independent experiments (transfection followed by titration) is shown.