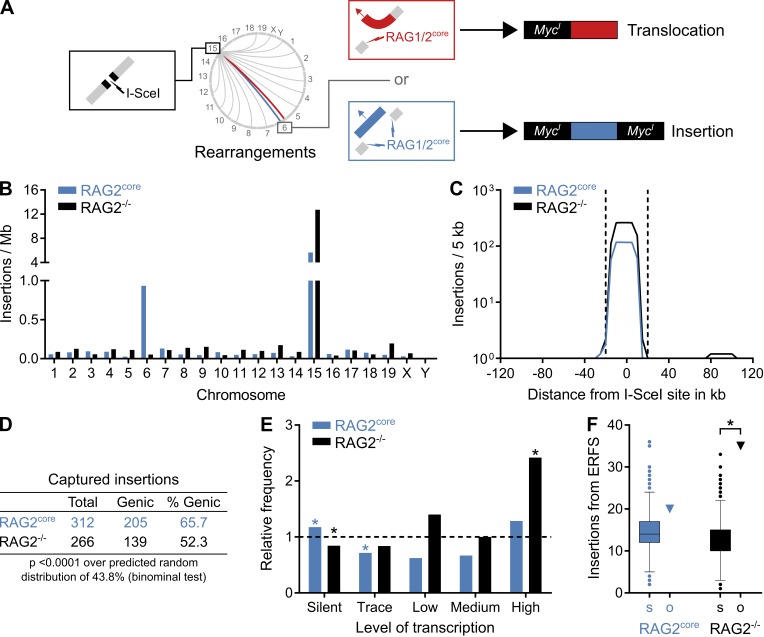
Figure 4.
Landscape of insertions in primary pro–B cells by TC-Seq. (A) Cartoon diagram comparing RAG1/2core-induced translocations and insertions. In a translocation (red), RAG1/2core introduces a single DNA break (red lightning) that recombines with the cleaved I-SceI site at MycI (black lightning) on chromosome 15. The resulting translocation contains MycI only on one side. In an insertion (blue), RAG1/2core causes tandem DNA breaks (blue lightning), thereby excising a DNA fragment that subsequently reintegrates into the cleaved I-SceI site. The resulting insertion is flanked by MycI on both sides. (B) Origin of insertions by chromosome. Events were normalized per megabase to account for different chromosome sizes. (C) Profile of insertions near the I-SceI site in 5-kb intervals. Dashed lines indicate the ±20-kb region excluded from the analysis for D–F because of saturation. (D) Proportion of insertions from genic regions. (E) Frequency of insertions derived from differentially transcribed genes compared with a random model (dashed line). Asterisks indicate values significantly different from random (P < 0.01, binominal test). (F) Observed number of insertions (o, triangle) originating from ERFSs compared with the random Monte Carlo simulation (s, Tukey boxplot). Asterisks indicate significant enrichment (P < 0.0001, binominal test). For D–F, events from the saturated I-SceI region, cryptic I-SceI sites, and other portions of the genome were excluded (see Materials and methods). Data analysis was performed with pooled RAG2core and RAG2−/− TC-Seq libraries (two independent experiments each).