Abstract
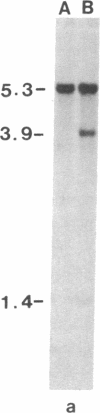
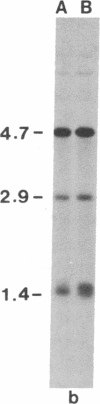
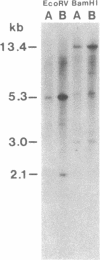
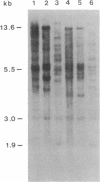
Restriction site variation in DNA that encodes rRNA (rDNA) was surveyed among 714 offspring within 31 lineages (26 genotypes) of obligate asexually reproducing Taraxacum officinale (dandelions). Although clonal offspring are expected, plants with nonparental rDNA were produced from two parents that were themselves siblings (same genotype). The variation is best characterized by the loss of an EcoRI restriction site that maps to the spacer region in the parental rDNA and is most likely involved in amplification of rare or unique rDNA repeats. In one family, 41 surveyed offspring lacked the EcoRI site. In the other family, only 1 of 26 offspring lost the EcoRI site. Other classes of DNA surveyed, chloroplast DNA and the alcohol dehydrogenase 2 gene (Adh2), showed no variation. However, offspring with nonparental rDNA also had nonparental alcohol dehydrogenase 1 (Adh1) restriction fragments. Because somatic mutations in plants can be incorporated into reproductive tissue, we propose that somatic events affecting at least both multicopy rDNA and DNA homologous to the maize Adh1 gene occurred at different developmental times in the two families. An event early in development would result in all variant offspring; an event late in development would result in a single variant offspring. These results support the view that mutation (in the broad sense) influences the level of genotypic variation in asexual organisms, which may facilitate adaptive evolution of asexual species.
Full text
PDF
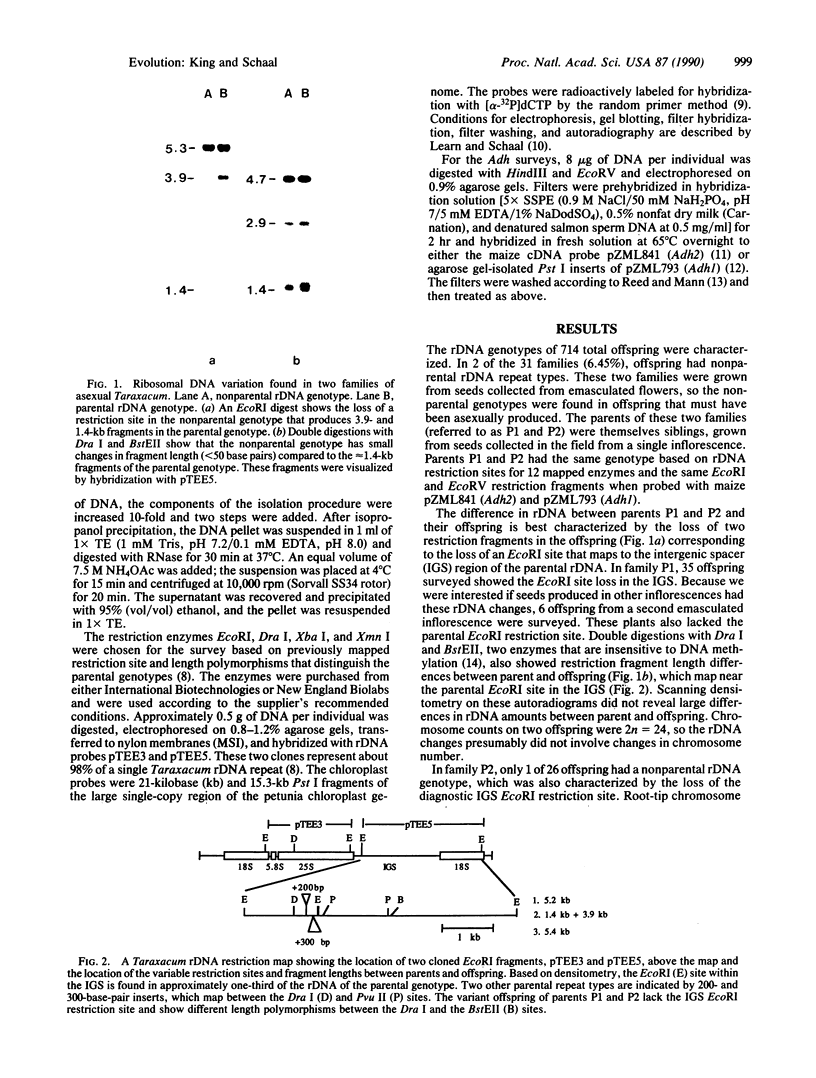
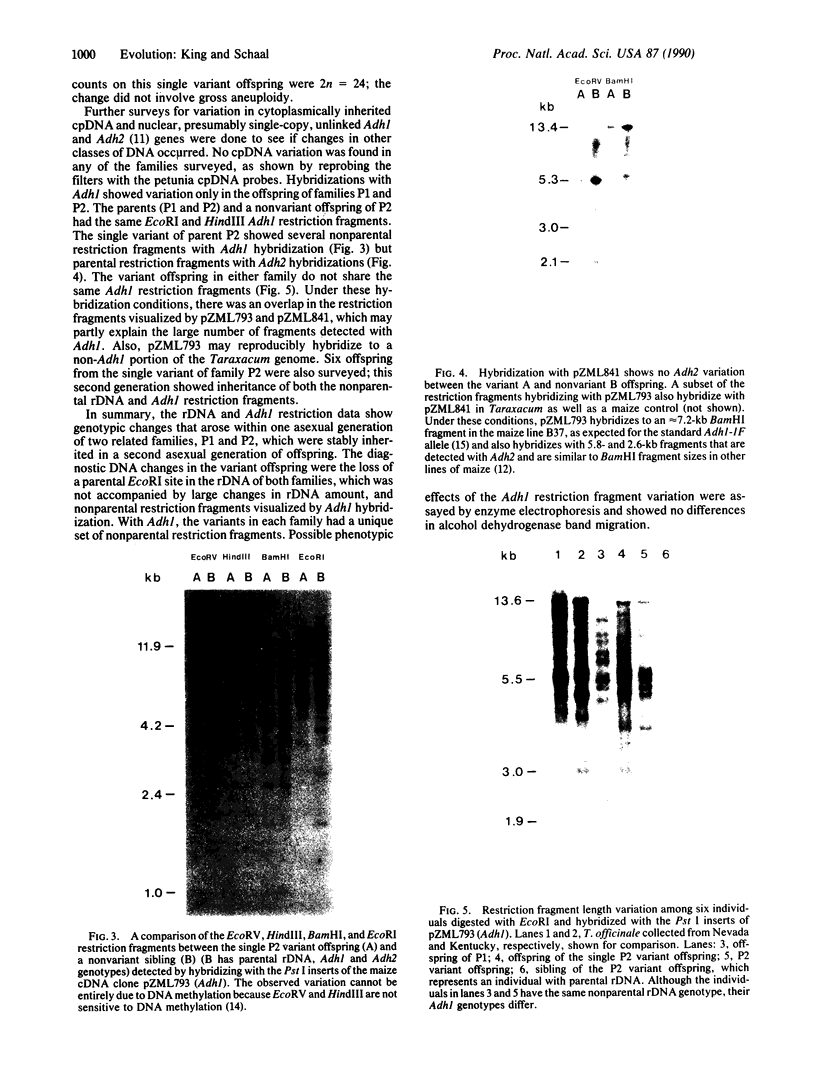


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bostock C. J. Mechanisms of DNA sequence amplification and their evolutionary consequences. Philos Trans R Soc Lond B Biol Sci. 1986 Jan 29;312(1154):261–273. doi: 10.1098/rstb.1986.0006. [DOI] [PubMed] [Google Scholar]
- Dennis E. S., Gerlach W. L., Pryor A. J., Bennetzen J. L., Inglis A., Llewellyn D., Sachs M. M., Ferl R. J., Peacock W. J. Molecular analysis of the alcohol dehydrogenase (Adh1) gene of maize. Nucleic Acids Res. 1984 May 11;12(9):3983–4000. doi: 10.1093/nar/12.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E. S., Sachs M. M., Gerlach W. L., Finnegan E. J., Peacock W. J. Molecular analysis of the alcohol dehydrogenase 2 (Adh2) gene of maize. Nucleic Acids Res. 1985 Feb 11;13(3):727–743. doi: 10.1093/nar/13.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorák J., Appels R. Investigation of Homologous Crossing over and Sister Chromatid Exchange in the Wheat Nor-B2 Locus Coding for Rrna and Gli-B2 Locus Coding for Gliadins. Genetics. 1986 Aug;113(4):1037–1056. doi: 10.1093/genetics/113.4.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Ono A., Ueda T. Synthesis of decadeoxyribonucleotides containing N6-methyladenine, N4-methylcytosine, and 5-methylcytosine: recognition and cleavage by restriction endonucleases (nucleosides and nucleotides part 74). Nucleic Acids Res. 1987 Jan 12;15(1):219–232. doi: 10.1093/nar/15.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs M. M., Dennis E. S., Gerlach W. L., Peacock W. J. Two Alleles of Maize ALCOHOL DEHYDROGENASE 1 Have 3' Structural and Poly(a) Addition Polymorphisms. Genetics. 1986 Jun;113(2):449–467. doi: 10.1093/genetics/113.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984 Dec;81(24):8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980 Apr 3;284(5755):426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- Taira T., Kato A., Tanifuji S. Difference between two major size classes of carrot rDNA repeating units is due to reiteration of sequences of about 460 bp in the large spacer. Mol Gen Genet. 1988 Jul;213(1):170–174. doi: 10.1007/BF00333416. [DOI] [PubMed] [Google Scholar]
- Tartof K. D. Unequal mitotic sister chromatin exchange as the mechanism of ribosomal RNA gene magnification. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1272–1276. doi: 10.1073/pnas.71.4.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]