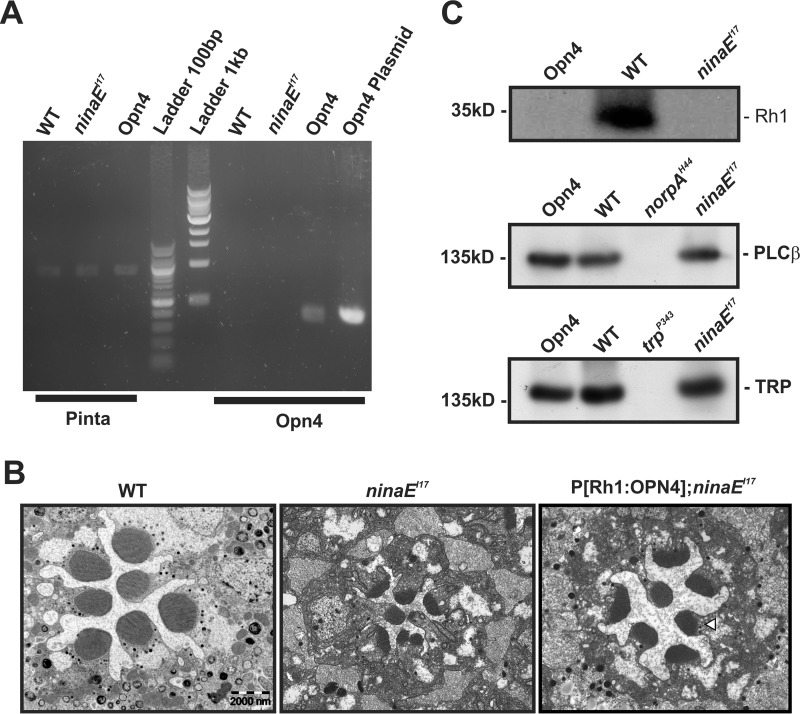
FIGURE 1.
Characterization of Drosophila transgene expressing mouse melanopsin (OPN4). A, Opn4 mRNA was detected in P[Rh1:OPN4];ninaEI17 transgenic flies. RT-PCR analysis, using primers of Opn4, showed expression of Opn4 mRNA (predicted size, 356 bp) in fly heads of opn4;ninaEI17 transgenic flies but not in the heads of WT or ninaEI17 mutant flies. An OPN4 plasmid was used as a positive control for the Opn4 primers. Primers for pinta (“prolonged depolarization after-potential is not apparent” (31); predicted size, 878 bp) were used as a positive control for cDNA synthesis. The middle lanes show the 100-bp and 1-kb ladder. The presented RT-PCR represents four independent experiments. B, expression of OPN4 partially rescued ninaEI17 retinal structural deformation. Representative EM of freshly eclosed opn4;ninaEI17 retina showing partial prevention of shrinkage of their rhabdomeres relative to ninaEI17 mutant and WT flies. Thin EM sections of ommatidia from ninaEI17 mutant (middle column), WT (left column), and opn4;ninaEI17 transgenic flies (right column) are shown. The white arrowhead indicates the location of the rhabdomeral stalk in which OPN4 is localized (see Fig. 2). Scale bar, 2 μm for all panels. C, Western blotting analysis showing no expression of Rh1 photopigment in opn4;ninaEI17 and ninaEI17 heads but normal expression of PLCβ and TRP. Top, Rh1 appeared only in WT and not in opn4;ninaEI17 and ninaEI17 flies. Middle, normal level of PLCβ appeared in opn4;ninaEI17 and ninaEI17 flies but not in the norpAH44 null PLCβ mutant (40) (negative control). Bottom, normal level of the TRP channel appeared in opn4;ninaEI17 and ninaEI17 flies but not in the trpP343 null mutant (67) (negative control). The labeling intensity of αRh1, αGqα, αPLCβ, αTRP, αTRPL, and αdMoesin (protein loading control; see “Experimental Procedures”) was compared between WT, opn4;ninaEI17, and ninaEI17 (n = 3). The figure shows expression levels of several signaling proteins, which have strong effects on the Drosophila LIC when their expression levels are reduced (i.e. Gqα is not presented because its level has to be reduced to <30% of normal to have a detectable effect on the LIC (50).