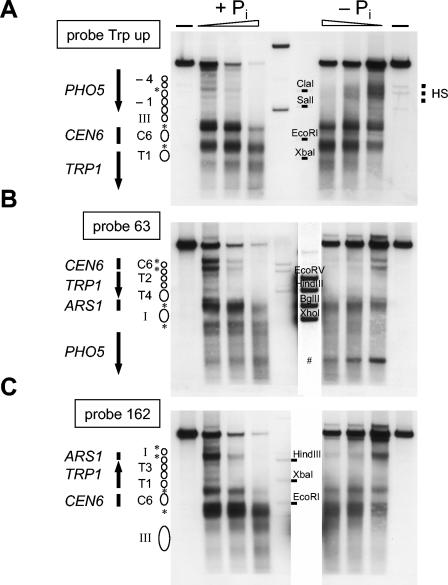
FIG. 5.
The chromatin structure of regions adjacent to the PHO5 promoter on plasmid pTAP5C remain largely unchanged upon induction. Nuclei of strain YS18018 carrying pTAP5C and grown under repressive (+Pi) or inducing (−Pi) conditions were analyzed by DNase I digestion and indirect end labeling (wedges and dashes, as in Fig. 2A). HindIII was used for secondary cleavage in panel A and ClaI in panels B and C, and probes were as indicated on the left of the blots (see also Fig. 1B). The relevant genetic regions as well as schematics of the chromatin structure (labeled and stippled circles, asterisks, and dashed lines for the extended hypersensitive site HS, as in Fig. 1A and B) are indicated at the sides of the blot. The blot of panel A was also probed with probe Trp down with the same result as in the “Plasmid” panel in Fig. 2A (P. Korber and D. Blaschke, data not shown). The two middle lanes of all blots contain markers for different probes. Only one of the two lanes is relevant for a particular probe and is labeled accordingly. The marker lane for probe Trp up (A) was underloaded and the position of the restriction fragments as indicated by dashes was deduced from a longer exposure. Panels B and C show the same blot which was probed with two different probes. The marker lane for probe 63 (B) was overloaded; therefore, a shorter exposure of this lane is shown in panel B and it was cut out in panel C. The pound sign in panel B denotes an artifact band.