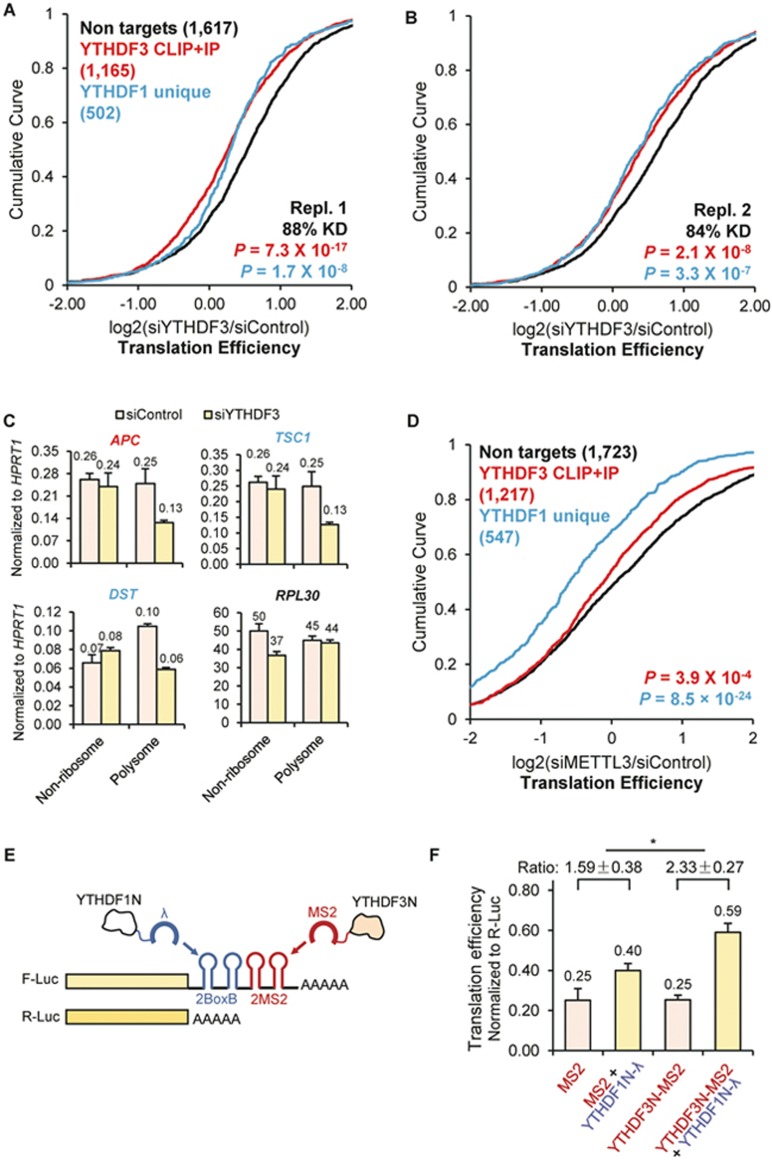
Figure 2.
YTHDF3 promotes translation efficiency of its mRNA targets and facilitates the function of YTHDF1. (A-B, D) Cumulative distribution of log2-fold changes of translation efficiency (ratio between ribosome-bound fragments and input RNA) between siControl and siYTHDF3, biological replicate 1 (A), siYTHDF3, biological replicate 2 (B), and siMETTL3 (D). Three groups of genes were plotted: non-targets (neither targets of YTHDF1 nor targets of YTHDF3, black), YTHDF3 CLIP+IP (high-confidence YTHDF3 targets, red), and YTHDF1 unique (YTHDF1 targets that are not targets of YTHDF3, blue). Number of genes in each group was indicated in parentheses. P values were calculated from a two-sided Mann-Whitney test compared to non-targets. (C) Redistribution of representative targets in non-ribosome and polysome portions of mRNPs upon depletion of YTHDF3 measured by RT-qPCR. APC, a YTHDF3 target; TSC1 and DST, YTHDF1 unique targets; and RPL30, a non-target. Error bars, mean ± sd, n = 2, technical replicates. (E) Construct of the double tethering assay. A sequence of two BoxB followed by two MS2 stem loops was inserted at the 3′UTR of F-Luc (firefly luciferase) mRNA. The C-terminal YTH domains of YTHDF1 and YTHDF3 were respectively replaced with λ peptide (binding BoxB motif) and MS2 binding protein (binding MS2 motif). R-Luc (renilla luciferase) mRNA was used as an internal control for normalizing luciferase signals from different samples. (F) Translation efficiency of F-Luc normalized with R-Luc 4-hour post F-Luc induction, with the expression of effectors indicated at x-axis. The ratio between YTHDF1N-λ (yellow) and the corresponding control sample (grey) was calculated. Error bars, mean ± sd, n = 3. P = 0.05 (paired two-sided Student's t-test).