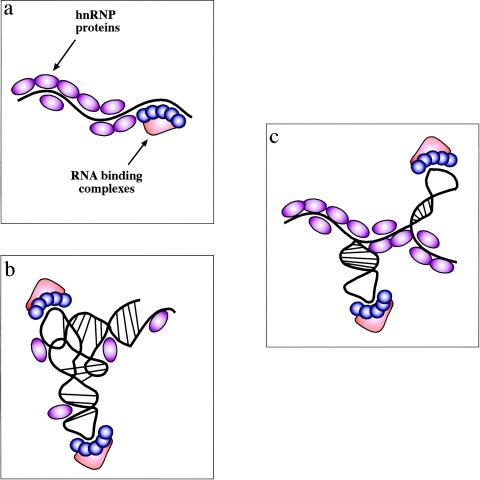
FIG. 1.
Experimental models of RNA secondary structures in mRNAs. There are three possible experimental models of RNA secondary structures in mRNAs. (a) The first one is represented by the view that hnRNP proteins bind the mRNA as it gets transcribed by RNA polymerase II and keep it in a largely linear conformation. In this case, binding of specific factors is regulated only by the competitive advantage provided by sequence-specific interactions over the generic RNA binding affinities of all hnRNP proteins. (b) The opposite situation is one where the drive to form RNA secondary and tertiary structures is stronger than the ability of RNA-binding proteins to prevent it (and maybe even stabilized by these proteins). In this case, the role played by generic RNA-binding proteins is severely reduced and specific complexes can bind through a mix of sequence-specific and structure-specific recognition. (c) Between these two models is a situation that should encompass many cellular mRNAs. In this case, the potential “ironing” of the mRNA by its weak or aspecific interactions with hnRNPs can indeed maintain the mRNA in a largely linear conformation. However, in particular regions the mRNA is still able to form localized RNA structures which might represent, together with the nucleotide sequence, preferential binding sites for specific nuclear complexes. Of course, given the enormous variety of mRNAs produced by the cell these models cannot be considered mutually exclusive, although there is probably a distinct preference for the model in panel c.