FIG. 4.
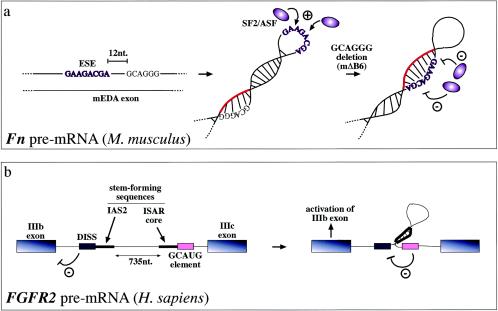
Steric hindrance has also been found to occur in ESE sequences, which promote recognition of the correct 3′ splice sites and 5′ splice sites. In the case of the mouse (and human) fibronectin EDA exons, secondary structural elements can stabilize the conformation of the ESE sequence and enhance its SR protein binding capabilities (13, 77). For example, mutations that do not directly affect the mouse ESE sequence have been demonstrated to cause a conformational change in this region (from a loop to a stem) which hinders SF2/ASF protein binding and in this way abolishes exon recognition (a). Highlighted in red is the RNA region which, following the mΔB6 deletion, blocks the ESE sequence. Alternatively, RNA secondary structures can also function on ESE/ESS regulatory regions indirectly. For example, in the FGFR2 gene they contribute to regulating the inclusion of the mutually exclusive IIIb (expressed in epithelial cells) and IIIc (expressed in mesenchymal cells) exons. In this case, the function of the stem structure formed by the intronic activating sequence 2 (IAS2) and intronic splicing activator and repressor (ISAR) element would be that of approximating an inhibitory intronic sequence (a GCAUG-rich sequence) relative to the distal intronic splicing silencer (DISS) element, which would normally repress exon IIIb inclusion (4). Inactivation of this element would then lead to activation of exon IIIb splicing in epithelial cells (b).