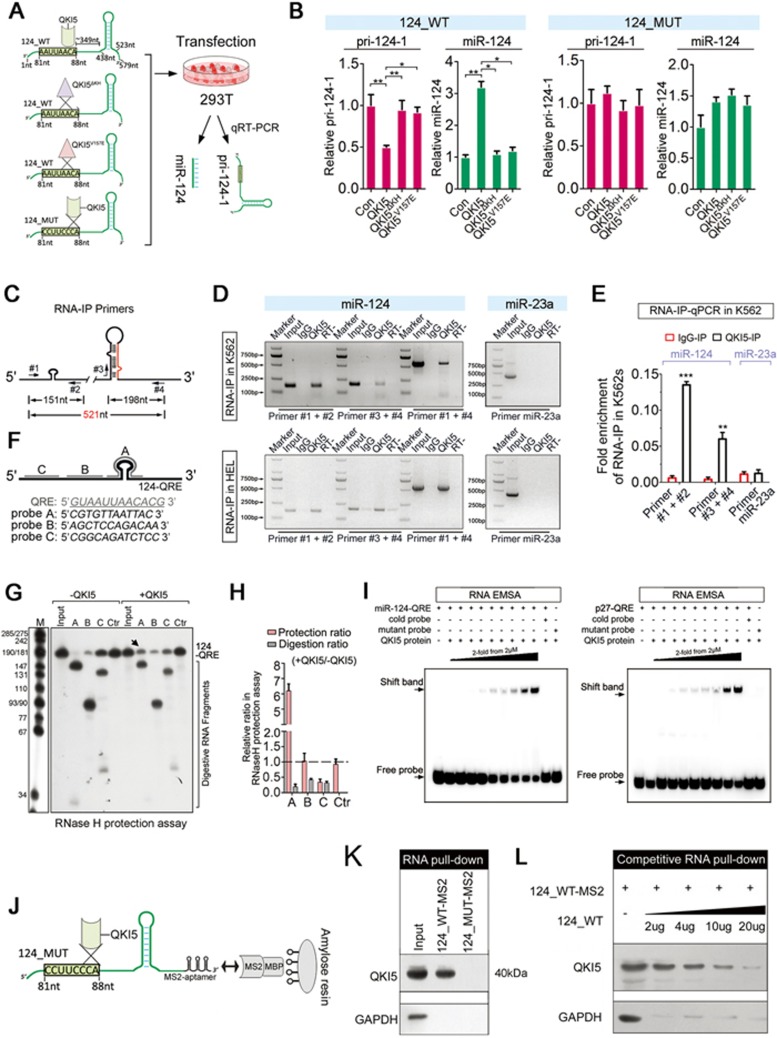
Figure 3.
QKI5 binds to the pri-124-1 transcript in vitro and in vivo. (A) A schematic representation of co-transfection with wild-type (124_WT) or mutant (124_MUT) pri-124-1 constructs and either wild-type (QKI5) or mutant (QKIΔKH and QKI5V157E) QKI5 constructs. The nucleotide positions of the QRE and stem loop are indicated in the cloned pri-124-1 sequence used in this study. (B) q-PCR shows the changes in the levels of pri-124-1 (red bar) and miR-124 (green bar) upon transfection of the indicated combinations into 293T cells as shown in A. (C) A schematic representation of the primers used in RNA-IP-PCR analysis. (D) RNA-IP-PCR assays for pri-124-1 performed on anti-QKI5 immunoprecipitates (QKI5) from K562 and HEL cell lysates. An unrelated IgG served as the negative control (IgG). RT: no reverse-transcribed PCR. QKI5 RNA-IP assays for the irrelevant pri-23a are shown in the right panel. (E) q-PCR analysis of pri-124-1 and pri-23a associated with QKI5 evaluated by RNA-IP assays in K562 cells. The RNA-IP-q-PCR results are shown as fold enrichment compared with input. (F) A schematic representation of the DNA oligonucleotides used in the RNase H protection assays. The antisense DNA oligonucleotides A-C span different regions of 124-QRE RNAs, whereas Ctr is an unrelated control oligonucleotide. (G, H) RNase H protection assays. RNase H cleavage results of reaction with either 124-QRE RNAs or QKI5-protected 124-QRE RNAs targeted by antisense DNA oligonucleotides A-C (lanes 2-4 and 7-9) or by an unrelated control oligonucleotide (Ctr, lanes 5 and 10; G). Additional control reactions were performed in the absence of oligonucleotide (input, lanes 1 and 6). Arrow indicates a reduced cutting site (G). The quantitative data were shown in H. The relative protection ratio was defined by the percentage of undigested 124-QRE in reactions with QKI5 protein to that without QKI5, and the relative digestion ratio was defined by the percentage of digested segments in reactions with QKI5 protein to that without QKI5. The data were quantified using ImageJ software. (I) In vitro association of QKI5 with the pri-124-1 QRE as identified by an RNA-EMSA assay in which 5′-biotin-labeled miR-124-QRE probes were incubated with different concentration of purified Flag-QKI5 (the left panel). A reported 5′-biotin-labeled p27-QRE probe was used as the positive control (the right panel). A mutant probe was used as the control, and the unlabeled miR-124-QRE probe was used in the competitive assays. (J) A schematic representation of the RNA pull-down assays using MS2-tagged pri-124-1 affinity purification. (K) Immunoblot of endogenous QKI5 in RNA pull-down assays from 293T cells transfected with either MS2-tagged wild-type pri-124-1 (124_WT-MS2) or MS2-tagged QRE mutant pri-124-1 (124_MUT-MS2) constructs. An unrelated protein GAPDH was used as the control. (L) The competitive association of non-MS2-tagged pri-124-1 with endogenous QKI5 in RNA pull-down assays performed on 293T cells co-transfected with 124_WT-MS2 and increased dosages of non-tagged 124_WT. Error bars reflect SEM from three biological replicates if not stated otherwise. Significance was determined by t-test with *P< 0.05; **P< 0.01; ***P< 0.001.