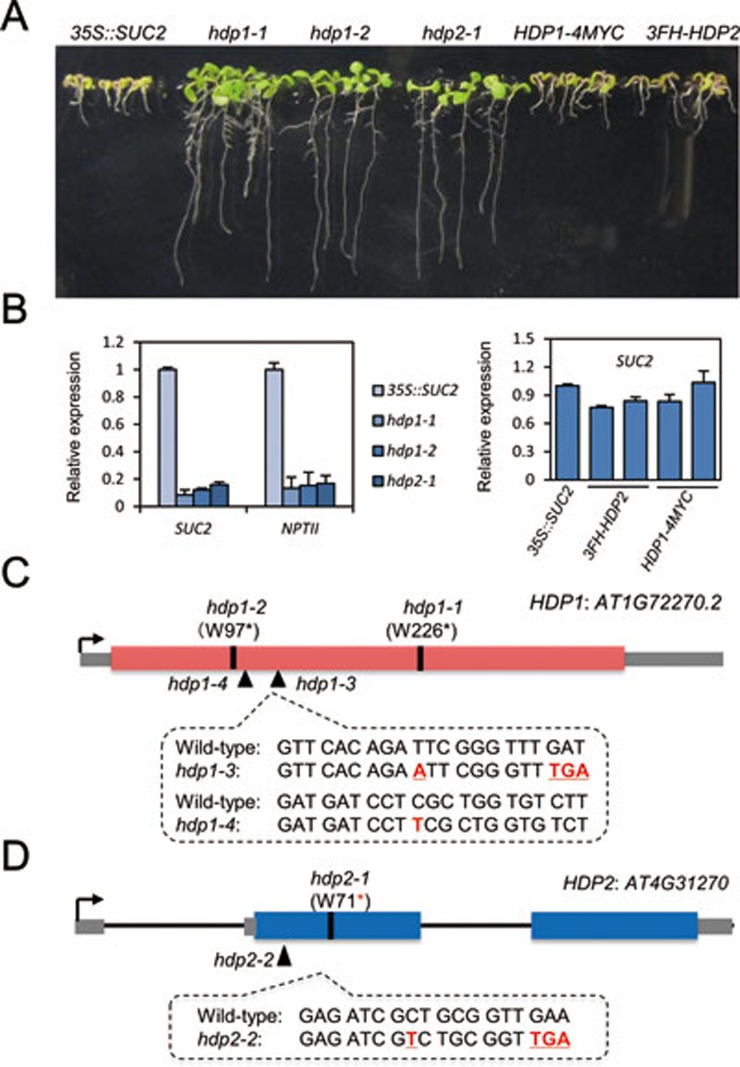
Figure 1.
HDP1 and HDP2 prevent transcriptional silencing of transgenes. (A) Identification of hdp1 and hdp2 mutants. Introduction of HDP1 and HDP2 genomic DNA fully complemented the root phenotype in hdp1-1 and hdp2-1 mutants. (B) RT-qPCR showing significantly reduced transcript levels of the transgenes SUC2 and NPTII in hdp1-1 and hdp2-1 mutants in comparison to 35S::SUC2 plants. HDP1 and HDP2 genomic DNA rescued the silencing of SUC2 transgene in hdp1-1 and hdp2-1 mutants. Two representative transgenic lines were selected and relative expression of SUC2 transcript was normalized to 35S::SUC2 plants. Three independent biological replicates were carried out for statistical analysis. See also Supplementary information, Figure S1. (C) Genomic structure and mutant alleles of HDP1 gene. For hdp1-1 and hdp1-2 mutants, nucleotide substitutions of “G” to “A” lead to the changes of 226th and 97th amino acids from tryptophan (W) to stop codons. hdp1-3 and hdp1-4 mutants are generated by CRISPR/Cas9-mediated genome editing. Nucleotide insertions of “A” and “T” were identified in hdp1-3 and hdp1-4 mutants, respectively. Red box indicates the coding region of AT1G72270 gene and gray box indicates untranslated regions. (D) Genomic structure and mutant alleles of HDP2 gene. For hdp2-1 mutant, a G-to-A substitution causes a pre-mature stop codon. An insertion of “T” was identified in hdp2-2 mutant. Blue box indicates the coding region of AT4G31270 gene and gray box indicates untranslated regions.