Fig. 7.
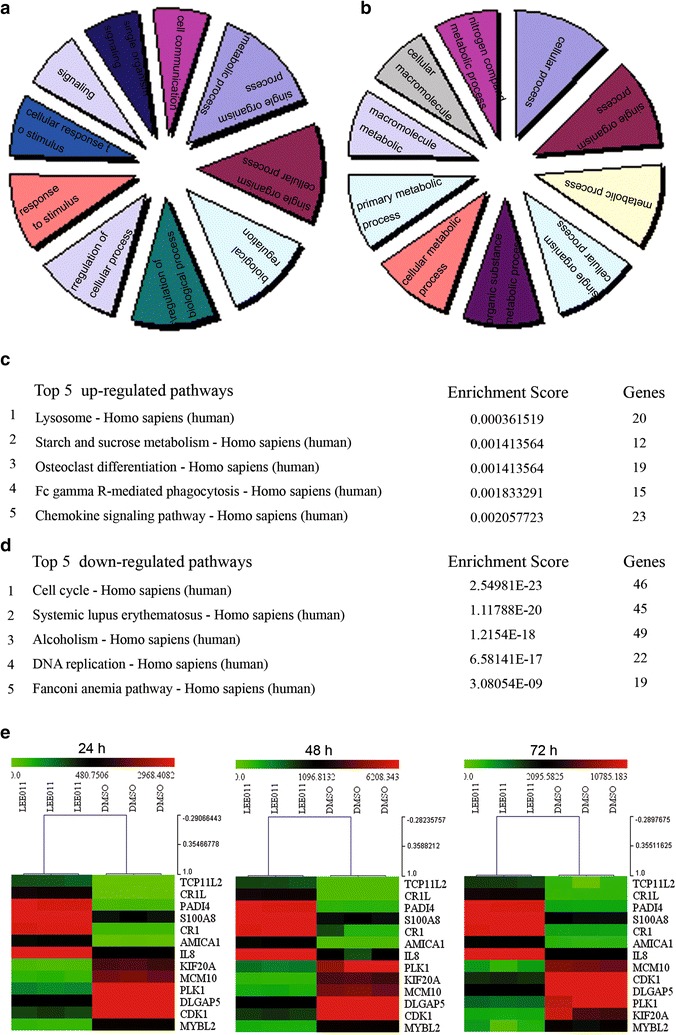
Gene ontology and KEGG pathway analysis of mRNA expression profiles in LEE011-treated HL-60 cells. a The most enriched GO terms for upregulated transcripts. b The most enriched GO terms for downregulated transcripts. c The top five enriched pathways for upregulated transcripts from KEGG pathway analysis. d The top five enriched pathways for downregulated transcripts from KEGG pathway analysis. The most enriched pathway was cell cycle, with a P value of 2.54981E−23. The cell cycle pathway included BUB1, BUB1B, BUB3, CCNA1, CCNA2, CCNB1, CCNB2, CCNE2 and CDC25A, amongst others. e Cluster analysis of several genes whose expression was detected by real-time PCR in HL-60 cells treated with 1uM LEE011 for 24, 48 and 72 h. Gene expression levels for each sample were normalized to the expression level of GAPDH within a given sample (−∆Ct). The relative expression of each gene was calculated using the equation: 106 × log2 (−∆Ct). Gene expression differences between the DMSO-treated and the LEE011-treated samples were analyzed using Multi Experiment View (MEV) cluster software
