Fig. 2.
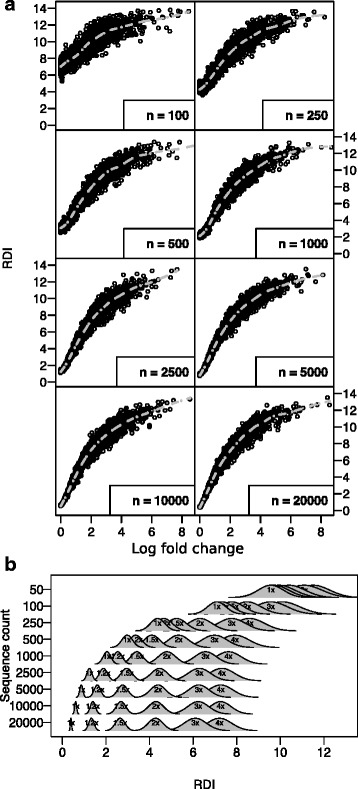
The RDI metric scales with differences in gene frequency. Simulated datasets were generated by randomly drawing genes from a set of fixed probability vectors. Probabilities were generated by perturbing a constant baseline probability vector such that the absolute log-fold difference in each gene was between 0 (no change) and 8 (256-fold increase or decrease in each gene) relative to baseline. Each perturbation vector was used to generate datasets containing varying numbers of sequences (n = 50 to 20,000), and a set of equally-sized baseline datasets were generated and compared to the perturbed datasets using the RDI metric. a The average RDI score for each perturbed dataset (y axis) is shown against the true average absolute log fold change (relative to baseline) of each perturbation vector (x axis). Spline models were fit to the data (dotted lines). b Mean and standard deviation of the RDI value was estimated from the spline model at multiple fold change values, and are plotted as probability density functions for a variety of different repertoire sizes (y axis)
